HRD is a common feature of high-grade serous ovarian cancer (HGSOC) as well as breast, prostate, and pancreatic cancers. Homologous recombination repair (HRR) is a DNA damage repair system responsible for the repair of DNA double-strand breaks. HRD may be used as a potential predictive biomarker for poly (ADP-ribose) polymerase 1 (PARP1) inhibition therapy response, based on its predicated synthetic lethality in the context of HRD-positive cells. HRD testing provides an opportunity to identify larger populations of cancer patients who may benefit from treatment with PARP inhibitors (PARPi).
What is HRD and why is it important in cancer development?
Cells respond to DNA damage by initiating multiple interconnected DNA repair pathways. HRR represents a central high-fidelity DNA damage repair system of DNA double-strand breaks. Functional defects in HRR are called HRD.1 The most known causes of HRD are loss of function mutations in genes involved in HRR including BRCA1, BRCA2, RAD51C, RAD51D, PALB2, as well as promoter hypermethylation of BRCA1.2 Over time, unrepaired or inaccurately repaired double-strand breaks lead to the accumulation of genomic aberrations, such as insertions and deletions, copy number alterations, or structural chromosomal rearrangements. These manifest as genomic instability, which drives carcinogenesis and disease progression.1
HRD is identifiable in many cancers. Approximately 15% of HGSOC harbor germline BRCA1/2 (gBRCA1/2) mutations. A total of 50% have HRD, conferred by either germline or somatic mutations in BRCA1/2 or mutations in other genes known to be involved in HRR.3 Similarly, gBRCA1/2 mutations are harbored by approximately 15% of triple negative breast cancers (TNBC) and a further 40% have HRD in the absence of gBRCA1/2 mutations. In advanced prostate cancers, 10%–12% of patients have germline or somatic BRCA2 inactivation and up to 25% contain a DNA damage repair defect.4 It remains to be determined whether HRD in different cancer types has the same biological and clinical implications.2
HRD testing provides an opportunity to identify larger populations of cancer patients who may benefit from treatment with PARP inhibitors (PARPi)
How does HRD predict sensitivity to PARPi?
PARPi are small molecule inhibitors of the PARP family of proteins.5 PARP1 is best known for its role in the base excision repair of single-strand DNA breaks.1 The discovery that single-agent PARP inhibition selectively killed BRCA-deficient cells was a breakthrough in exploiting synthetic lethality approaches in oncology (Figure 1).4,6
Pre-clinical evidence has shown that PARP can modulate the repair of double-strand DNA breaks, if cancer cells are HRD without the capacity to repair double-strand DNA breaks.7 PARPi trap the PARP1 protein on DNA at sites of single-strand DNA breaks. If trapped PARP1 is encountered by the DNA replication machinery, it leads to stalling of the replication fork, the collapse and the generation of a double-strand break, which cannot be repaired in HRD cells. Thus, PARPi prevent the DNA repair process, ultimately leading to the disruption of cellular homeostasis and cell death.8 HRD has been identified as predictive biomarker for PARPi therapy response in ovarian, and potentially in breast, and prostate cancers.9–11
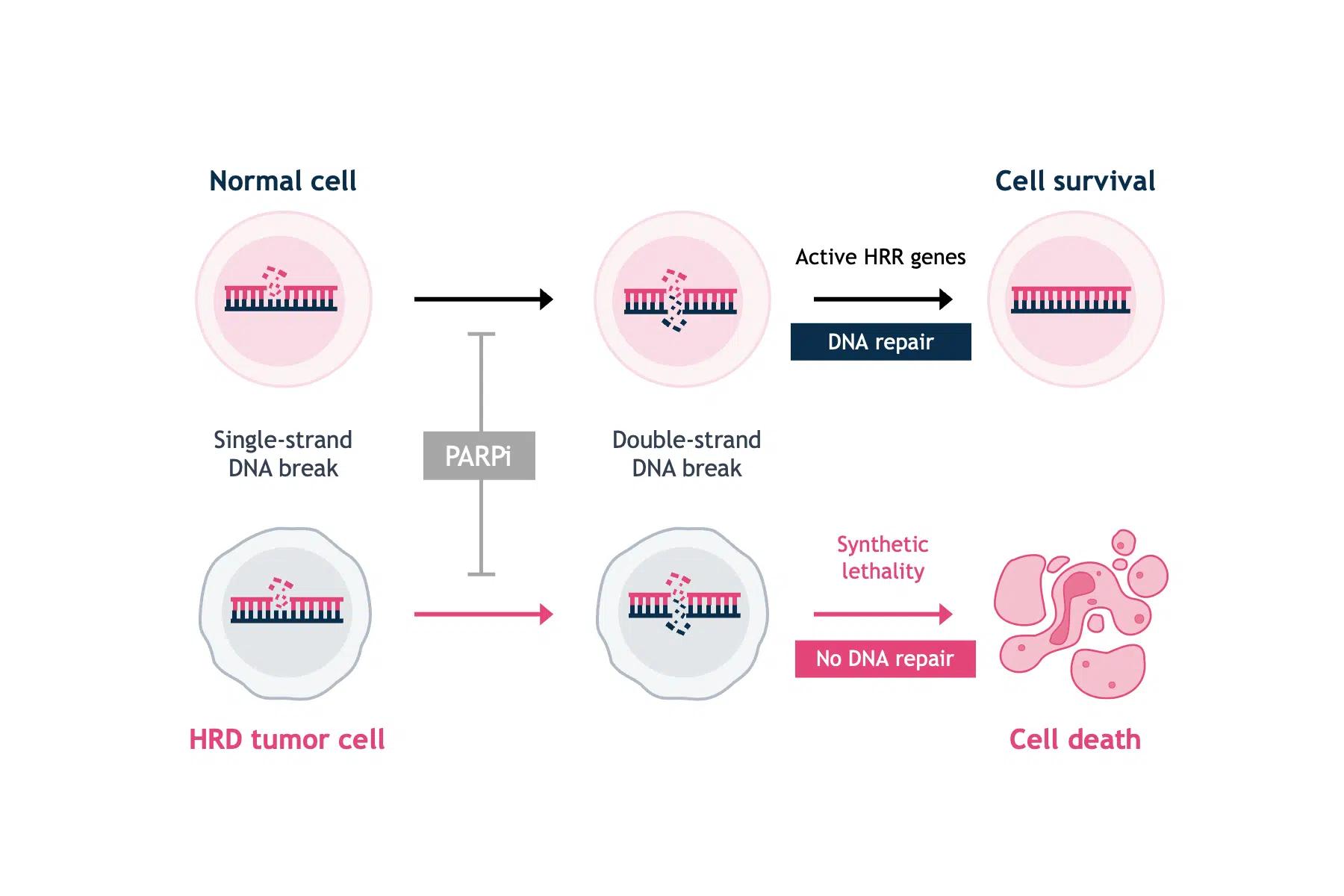
In the normal cell, PARP inhibition will cause DNA single-strand breaks to progress to double-strand breaks. However, functional HRR genes (e.g. BRCA1/2) will have the capability to repair DNA double-strand breaks and maintain cell viability.
In the tumorigenic cell, PARP inhibition will cause single-strand breaks to progress to double-strand breaks, but deficient levels of proteins that facilitate HRR (e.g. BRCA1/2) will decrease rates of double-strand breaks repair and allow accumulation of double-strand breaks—ultimately leading to cell death via apoptosis.
Assays for HRD detection in cancer – what are the challenges and perspectives?
The assessment of HRD is an important prognostic and predictive biomarker.1 Currently available testing methods for HRD fall into three categories: HRR gene-based tests, genomic scars and signatures, and functional assays.4
HRR gene-based tests
The most reliable clinical predictor of HRD and response to PARPi remains gBRCA mutations and FDA approval has been granted for gBRCA detection tests in ovarian cancer.12 However, the negative predictive value of BRCA mutation status is poor in the setting of platinum-sensitive relapsed HGSOC, with BRCA wild-type (BRCAwt) subgroups also deriving a significant, although numerically smaller benefit from PARPi. There are currently no approved diagnostic tests for HRD based on germline mutations of other HRR genes.4 Using germline mutations in HRR genes to predict HRD status has several drawbacks. Genetic testing often results in the detection of variants of uncertain significance where a particular rare genotype may not confer an HRD phenotype. Additionally, somatic reversion mutations in the BRCA genes can restore homologous recombination proficiency (HRP). HRDstatus is therefore a complex and dynamic phenotype.4
Genomic scars and signatures
Since a considerable number of genes with potentially variable biological impact are involved in HRR, elucidating the clinical implications of individual germline or somatic mutations in HRR coding genes is challenging. An alternative strategy is to identify the effects of deleterious mutations in HRR genes. The inability to repair DNA in tumors that have HRD results in genomic scars or mutational signatures that measure the patterns of somatic mutations that accumulate as a result of HRD, irrespective of the exact nature of the underlying etiology.1,4 Studies of genomic scars and signatures generally measure somatic copy number variation (CNV) and predict BRCA status through the quantification of large-scale transitions (LST), loss of heterozygosity (LOH) or the number of sub-chromosomal regions with allelic imbalance extending to the telomere (NtAI), combined in most tests to generate a genomic instability score.4 Current commercially available tests combine tumor BRCA mutation testing with a genomic instability score.
Functional assays
Conversely, functional HRD assays have the potential to provide a dynamic readout of the current HRR status. The most commonly used experimental system to estimate HRR has been to assess the amount of nuclear RAD51.4 RAD51 is a downstream protein in the HRRpathway that is loaded onto sites of DNA double-strand breaks to facilitate DNA strand invasion into the sister chromatid in cooperation with mediator protein complexes, especially BRCA1 and BRCA2 (Figure 2).1,4
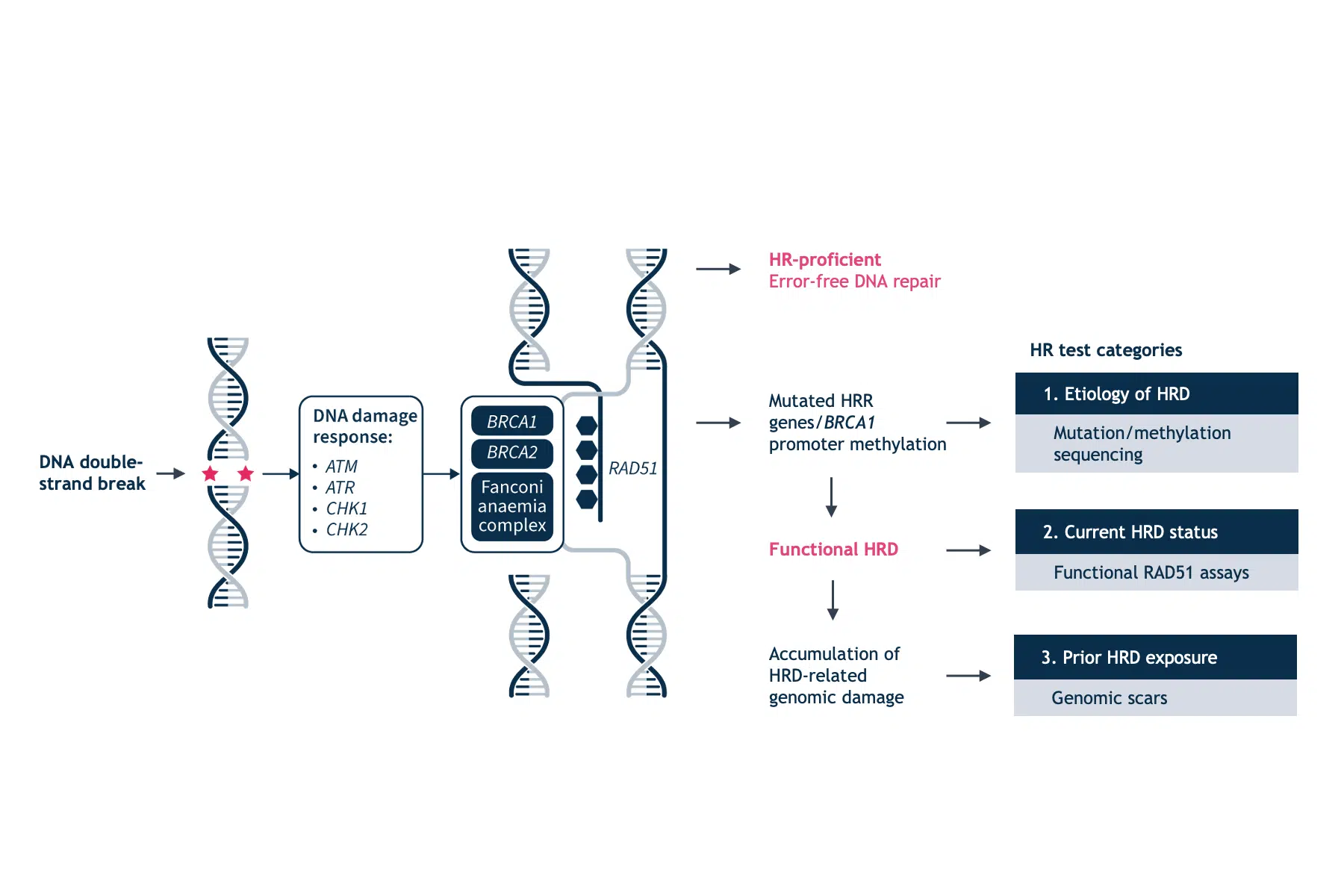
HRD cells are characterized by a reduction or inability to form RAD51 foci. This phenotype occurs irrespective of the underlying cause of HRD. In the presence of acquired reversion mutations that restore HRR, RAD51 foci are present. However, this technique also has its caveats. The RAD51 assays will not identify HRR defects downstream of RAD51 loading on to DNA and the RAD51 signal may be elicited by exogenous DNA damage. Another functional measure of HRD is the sensitivity to platinum salts, which in itself is a predictive marker for PARPi.5 Platinum sensitivity is a feature of HRD cells, and BRCA-mutant ovarian and breast tumors demonstrate increased platinum sensitivity.12 Since platinum-based therapies are a key component of first-line chemotherapy in ovarian cancer, prior platinum sensitivity has been evaluated as a surrogate clinical index for prediction of efficacy to PARP inhibition.12 However, there is still insufficient evidence to ascertain the clinical validity of functional assays in predicting PARP inhibition response.4
How can the SOPHiA DDM™ HRD Solution help accelerate HRD detection?
At SOPHiA GENETICS, we offer a novel approach to HRD detection with the SOPHiA DDMTM HRD Solution. In a single assay, our solution allows clinician researchers to:
- Identify HRR mutations through a targeted sequencing approach
- Measure genomic instability through a combination of low-pass whole genome sequencing and a powerful deep-learning algorithm
The comprehensive assay provides researchers with timely, in-house results on HRD status.
References
- Ngoi NYL, Tan DSP. ESMO Open 2021;6(3):100144.
- Gonzalez D, Stenzinger A. Genes Chromosomes Cancer 2021;60(5):299–302.
- Konstantinopoulos PA, Ceccaldi R, Shapiro GI, et al. Cancer Discov 2015;5:1137–54.
- Miller RE, Leary A, Scott CL, et al. Ann Oncol 2020;31(12):1606–22.
- Robinson D, Van Allen EM, Wu YM, et al. Cell 2015;161(5):1215–28.
- Neiger HE, Siegler EL, Shi Y. Int J Mol Sci 2021;22(11):5614.
- Huang X, Jan H, Xiao Q, et al. Front Oncol 2020;10:958.
- European Medical Journal. The Emerging Role and Value of Homologous Recombination Deficiency Testing in Advanced Ovarian Cancer: Interviews with Four Key Opinion Leaders. Available at: https://emj.emg-health.com/oncology/symposium/the-emerging-role-and-value-of-homologous-recombination-deficiency-testing-in-advanced-ovarian-cancer-interviews-with-four-key-opinion-leaders-s130721/. Accessed: February 2022.
- Robson ME, Tung N, Conte P, et al. Ann Oncol 2019;30(4):558–66.
- González-Martín A, Pothuri B, Vergote I, et al. N Engl J Med 2019;381(25):2391–402.
- de Bono J, Mateo J, Fizazi K, et al. N Engl J Med 2020;382(22):2091–102.
- Hoppe MM, Sundar R, Tan DSP, et al. J Natl Cancer Inst 2018;110(7):704–13.