Alamut™ Visual Plus
For variant annotation, deep exploration, and enhanced visualization
Variant annotation and interpretation on a genomic scale
Pinpointing pathogenic mutations from large, complex datasets can be difficult, time-consuming, and sometimes overwhelming. Alamut™ Visual Plus is a comprehensive, full genome browser for efficient and user-friendly variant interpretation.
Alamut™ Visual Plus, accessible through both SOPHiA DDM™ and as a stand-alone product, is used in renowned university medical centers, institutions, and genetics laboratories worldwide.
Highly appreciated by its users, the software accelerates the complex and time-consuming assessment of variants thanks to its user-friendly interface and integrated features for variant annotation and analysis.
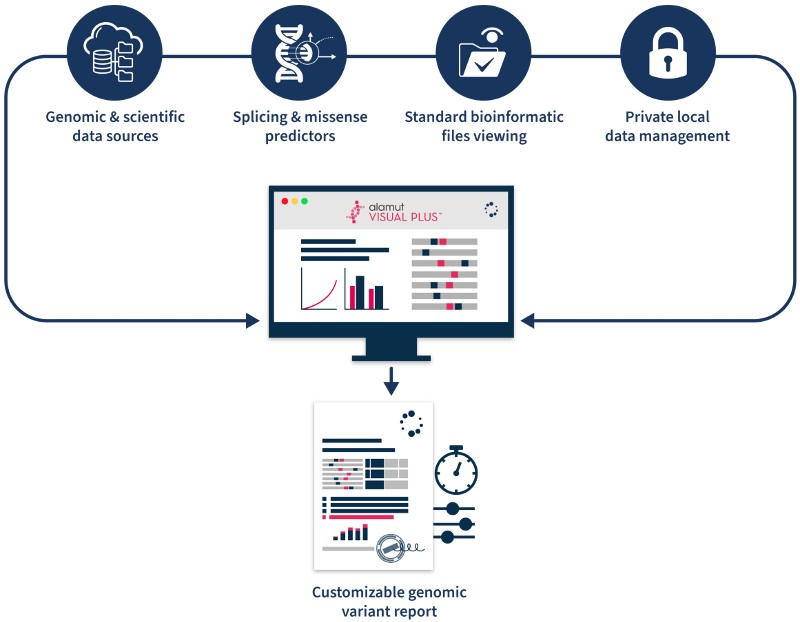
3 Key Features of Alamut™ Visual Plus for Variant Analysis
1. Comprehensive variant annotation
ACMG point-based classification with the option to filter based on evidence strength for each rule
International guidelines such as HGVS nomenclature
A single interface to access variant annotation information from >55 world-renowned curated databases and predictors such as ClinVar, dbSNP, BRIDGES, CADD, COSMIC, MedGen, Orphanet, dbVar, and DGV
High-quality in-silico missense and splice variant predictors such as REVEL, MaxEntScan, NNSPLICE, SIFT, and PolyPhen-2
Literature sources such as PubMed® and MasterMind®
2. Enhanced variant visualization
Comprehensive full-genome browser
GRCh37/38 assemblies and mitochondrial genome visualization
Structural variant track including copy number variation (CNV) analysis
Displays flanking regions and overlapping genes
BAM, VCF, BED, and Sanger file viewers
Simultaneous exploration of multiple genes
3. Efficient laboratory workflows
Direct link from SOPHiA DDM™
Private data management
Dynamic variant filtering
Customizable reporting
“I particularly like the visualization of GRCh37, GRCh38, or the mitochondrial genome, conveniently displaying regions close to the genes of interest and the option to see overlapping genes. Also, the option of knowing the impact of a variant on the overlapping gene, as the software enables analysis of several genes’ BAM sequences simultaneously”
Daniel Bellissimo, PhD
Director University Pittsburgh Medical Center Clinical Genomics Laboratory
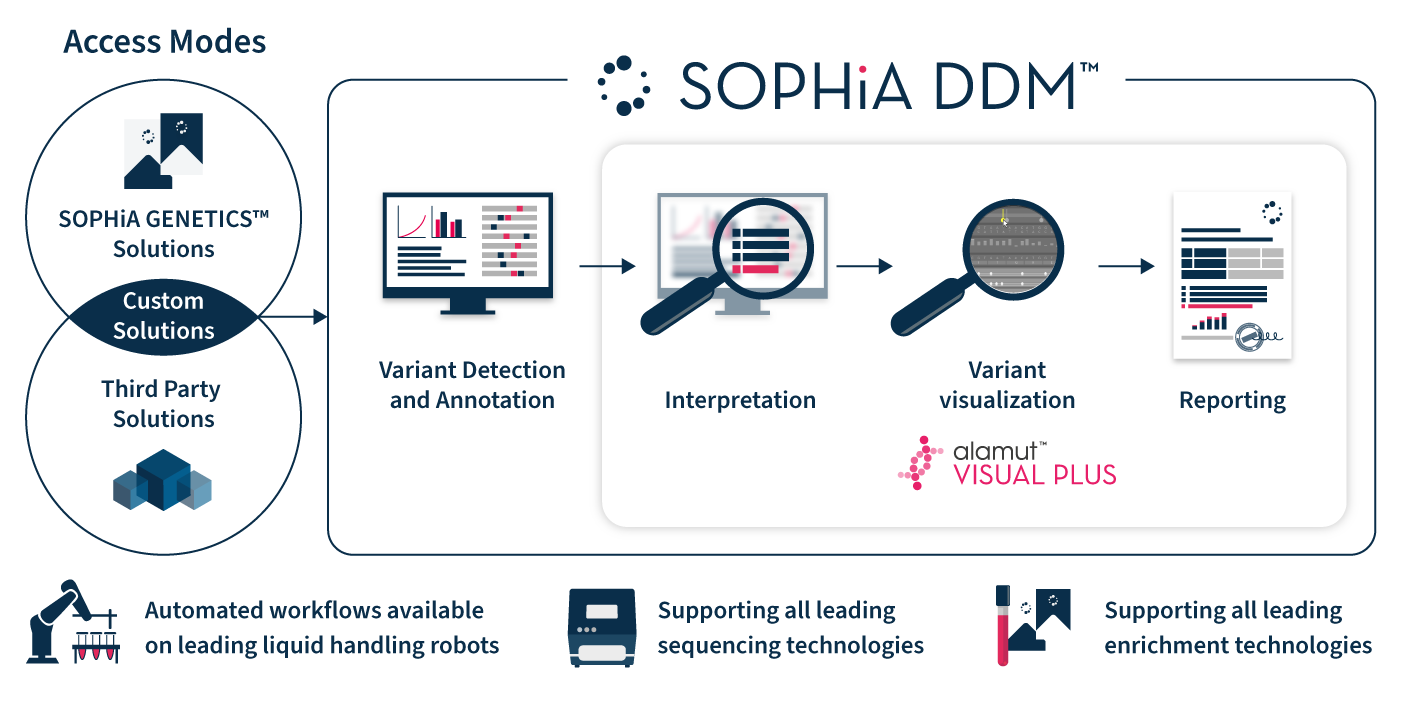
Alamut™ Visual Plus works in harmony with SOPHiA DDM™
to streamline any in-scope laboratory workflow
Resources
Application and technical notes
Related webinars
- Transcript analysis and splicing predictors - Dr Kai Lee Yap, Lurie Children's
- SOPHiA DDM™ and Alamut™ Visual Plus in action: Optimized variant prioritization for enhanced insights
- Integrating nuclear and mitochondrial DNA analysis to expand research into rare diseases
- R&D Collaboration with Professor Peter Devilee: The clinical significance of missense variants in breast cancer susceptibility genes – results from the BRIDGES project
Flyers
Related blogs and case studies
- Article Spotlight: Creating a splice variant decision tree
- Continuing to crack the mitochondrial genetic code
- Separating the wheat from the chaff: Overcoming the challenges of exome sequencing
- Motol University Hospital in Prague discover a novel cause of inherited cardiac disease with the SOPHiA DDM™ Clinical Exome Solution v3
Frequently Asked Questions
Try for free!
Get in touch to find out more.
Our client services team is on hand to answer any questions or set you up with a free trial.
If you’re an Alamut™ Visual Plus user with a technical query, please don’t hesitate to contact our dedicated technical support team.