The HGVS nomenclature guidelines are used worldwide for genetic variant interpretation but can seem complicated and difficult to understand and apply. That is why we have created this beginner’s guide to mutation nomenclature using the HGVS recommendations, with clear visual examples that break down the process into bitesize pieces.
1. What is HGVS nomenclature?
2. How to read mutation nomenclature: Breaking down the variant description
2.1 Reference sequence e.g., NM
2.2 Description of variant e.g., c.4375C>T
2.3 Predicted consequence e.g., p.(Arg1459*)
3. The 3 prime rule for mutation
4. Final thoughts and helpful tool
1. What is HGVS nomenclature?
The Human Genome Variation Society (HGVS) nomenclature standard was developed to prevent the misinterpretation of variants in DNA, RNA, and protein sequences. The HGVS nomenclature standard is used worldwide, especially in clinical diagnostics, and is authorized by the Human Genome Organisation (HUGO).1,2
HGVS General Terminology Recommendations1
| X Do not use | ✔️ Recommended terminology |
| Mutation or polymorphism | Variant, change, allelic variant Can be used for cancer tissue: Mutation load and tumor mutation burden |
| Pathogenic | Affects function, disease-associated, phenotype-associated |
HGVS follow recognized standards for the nomenclature of DNA and RNA nucleotides, the genetic code, amino acid descriptions, and cytogenetic band position in chromosomes.3
2. How to read mutation nomenclature: Breaking down the variant description
The HGVS recommendations for mutation nomenclature state that the format of a complete variant description should first include the reference sequence, followed by the variant description, and then the predicted consequence in parentheses. For example, NM-004006.2:c.4375C>T p.(Arg1459*) (Figure 1).
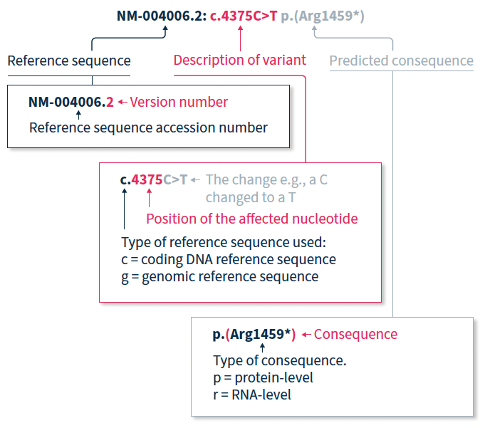
2.1. How to read mutation nomenclature: Reference Sequence
The HGVS nomenclature recommendations for sequence variants state that a complete variant description should begin with the reference sequence.1 The reference sequence accession number begins with a two-letter abbreviation (explained in Table 1), followed by a multi-digit number, and finally a version number.
Table 1. Meaning of the two-letter abbreviation at the beginning of a reference sequence accession number.
| Abbreviation | Reference sequence based on a: |
| NC | Chromosome |
| NG | Gene or genomic region |
| LRG | Locus Reference Genomic sequence: Gene or genomic region, used in a diagnostic setting |
| NM | Protein-coding RNA (mRNA) |
| NR | Non-protein-coding RNA |
| NP | Protein (amino acid) sequence |
2.2. How to read mutation nomenclature: Description of variant
The variant description begins by depicting the type of reference sequence used (c = coding DNA sequence, g = genomic reference sequence). When a protein-coding reference sequence is used (c), the nucleotide numbering begins with a 1, which represents the first position in the protein-coding region (the A of the translation-initiating ATG), and ends at the last position of the stop codon. Thus, if you divide the position number by 3, you can identify the affected amino acid in the protein sequence e.g., using the same example as above, 4375/3 = 1459, indicating that the predicted consequence affects amino acid 1459, which is an arginine. Different variants are indicated using different notations (explained in Table 2).
Table 2. HGVS notation and examples for the most common types of mutations2
| Notation | Example | Explanation |
| > | c.4375C>T | Substitution of the C nucleotide at position c.4375 with a T |
| del | c.4375_4379del or c.4375_4379delCGATT | Nucleotides from position c.4375 to c.4379 deleted |
| dup | c.4375_4385dup or c.4375_4385dupCGATTATTCCA | Nucleotides from position c.4375 to c.4385 duplicated |
| ins | c.4375_4376insACCT | ACCT inserted between positions c.4375 and c.4376 |
| delins | c.4375_4376delinsACTT or c.4375_4376delCGinsAGTT | Nucleotides from position c.4375 to c.4376 (CG) are deleted and replaced by ACTT |
2.3. How to read mutation nomenclature: Predicted consequence
When only DNA has been analyzed, the RNA- and protein-level consequences of the variant can only be predicted, and should thus be reported in parentheses e.g., p.(Arg1459*) is the predicted effect at protein level (p) for the example described above.
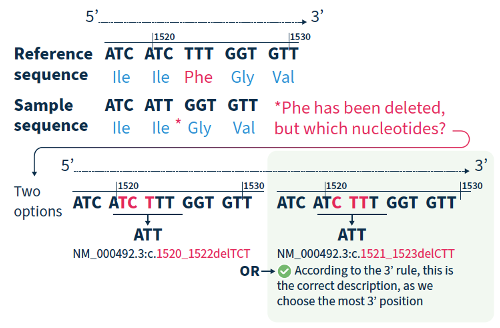
3. The 3 prime rule for mutation nomenclature
For all variant descriptions using HGVS nomenclature, the nucleotide at the most 3’ position of the variation in the reference sequence is arbitrarily assigned to have changed (see how to apply this rule in Figure 2).4 The exception is for deletions/duplications around exon junctions for which shifting the variant 3’ would place it in the next exon.5
4. Final thoughts and helpful tool
Although the HGVS recommendations can be difficult to understand and might take a bit of getting used to, if you break them down and refer to the examples in this guide, you are on the road to success!
If you want to accelerate your variant annotation and interpretation, Alamut™ Visual Plus is a comprehensive, full genome browser for efficient and user-friendly variant interpretation. The software accelerates the complex and time-consuming assessment of variants thanks to its user-friendly interface and integrated features for variant annotation and analysis.
Find out how Alamut™ Visual Plus applies the HGVS nomenclature recommendations to ensure that variant annotation follows the universally applied standards for variant analysis, interpretation, and reporting in our dedicated Technical Note.
Alamut™️ Visual Plus is for Research Use Only. Not for use in diagnostic procedures.
References
- https://varnomen.hgvs.org/bg-material/basics/
- http://varnomen.hgvs.org/bg-material/simple/
- http://varnomen.hgvs.org/bg-material/standards/
- http://varnomen.hgvs.org/recommendations/general/
- http://varnomen.hgvs.org/bg-material/numbering/#DNAc