Case Study – Motol University Hospital
Motol University Hospital in Prague discover a novel cause of inherited cardiac disease with the SOPHiA DDM™ Clinical Exome Solution v3
Country
Czech Republic
Research Challenge
Cardiac disease
Solution
SOPHiA DDM™ Clinical Exome Solution v3
The Institute of Biology and Medical Genetics at Motol University Hospital in Prague is a research centre for the analysis of hereditary diseases, part of the largest medical facility in the Czech Republic. The Genetics laboratory has a particular interest in understanding the genetic causes of cardiac diseases.
Research Challenge
The lab received a sample from a man who had experienced a cardiac event that led to cardiopulmonary resuscitation (CPR). To research the underlying genetic cause, the lab initially conducted next-generation sequencing (NGS) with a targeted panel comprised of 100 genes implicated in cardiac disorders. The SOPHiA DDM™ Platform was used to align and call variants from the raw NGS data and subsequently analyze and interpret the results. A potential causative variant was identified, but as a variant of uncertain significance, it required further research.
Confirmation
The lab chose to corroborate their findings using the SOPHiA DDM™ Clinical Exome Solution v3 (CES v3), which enables the analysis of 5000 clinically relevant nuclear and mitochondrial genes, including additional genes implicated in cardiac disease.
Results
The adoption of CES v3 successfully helped to corroborate the pathogenic variant: NM_001256850.1(TTN):c.9358_9610del p.(Arg3120Glufs*58), confirmed by Sanger sequencing. The lab used Alamut™ Visual Plus to visualize the pathogenic variant in the context of the rest of the genome (Figure). First, they highlighted the area of the CNV on the genome viewer and then looked at the gnomAD and ClinVar tracks for similar variants. A variant was absent in healthy controls in gnomAD, therefore, suspected to be causative. The variant had not been previously reported in ClinVar, or the literature and so was considered a novel variant.
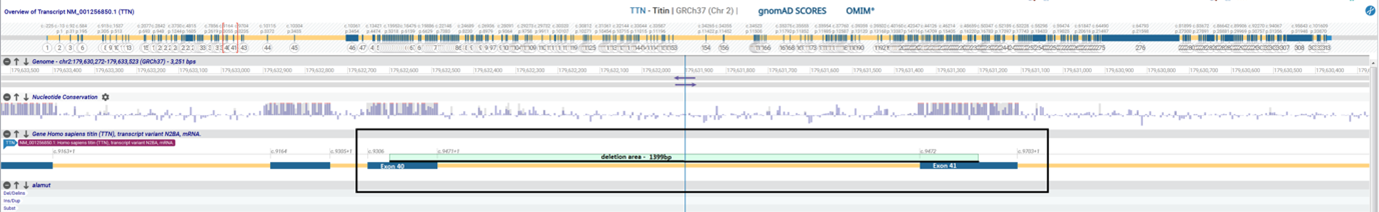
Visualization of the pathogenic variant in TTN in Alamut™ Visual Plus.
The detected CNV caused a frameshift and a premature termination codon 58 amino acids downstream, predicted to lead to a truncated or absent titin/connectin protein due to nonsense-mediated mRNA decay. Titin, or connectin, is a giant muscle protein expressed in the cardiac and skeletal muscles. It spans half of the sarcomere and plays a key role in muscle assembly, force transmission, and maintenance of resting tension. Mutations in TTN have previously been associated with cardiomyopathy and muscular dystrophy.
A segregation study found that although the proband’s healthy son lacked the pathogenic variant in TTN, his daughter did possess it and presented with arrhythmogenic right ventricular dysplasia.
Overall, the SOPHiA DDM™ Platform complemented by Alamut™ Visual Plus identified a novel pathogenic variant previously unreported in ClinVar and allowed a segregation study of the proband’s family for the TTN mutation.
“Community Frequency is a very useful tool that enables us to make a quick decision about whether or not to consider a variant and continue with a deeper inspection.”
Motol Hospital, Charles University,
Prague, Czech Republic









