Analyze HRD beyond BRCA by exploring multiple HRR genes simultaneously and potentially identify more samples sensitive to PARPi.
Genetic abnormalities in homologous recombination repair (HRR) genes, beyond BRCA1 and BRCA2, can lead to homologous recombination deficiency (HRD) in several cancer types, including ovarian, breast, prostate, and pancreatic cancers 1.
Focus on extended HRR sequencing in your research laboratory to increase the probability of identifying positive samples.
Sequence more genes beyond BRCA 1 and BRCA 2 to identify the cause of HRD associated with various cancers
Have your library ready to sequence in 1.5 days
Rely on high accuracy and coverage uniformity across the panel
Detect gene amplifications alongside single nucleotide variants (SNVs) and Indels with our proprietary algorithms
Accelerate interpretation with the SOPHiA DDM™ intuitive variant filters
Take advantage of guideline-driven variant pathogenicity ranking using machine learning
Use OncoPortal™ Plus to classify variants according to evidence-based annotations from the JAX-CKB database
Mutated HRR genes disrupt efficient DNA-repair thereby leading to genome-wide scarring known as HRD phenotype.
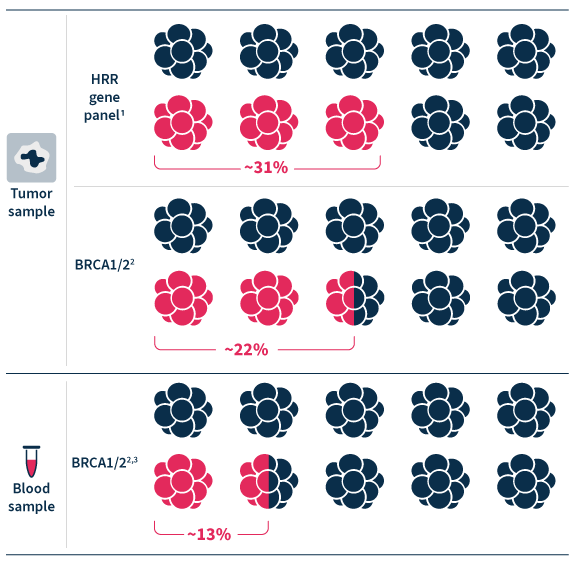
Increase the probability of identifying HRR mutations beyond BRCA1 and BRCA 2 by sequencing 28 HRR genes with SOPHiA DDM™ Extended Homologous Recombination Solution.
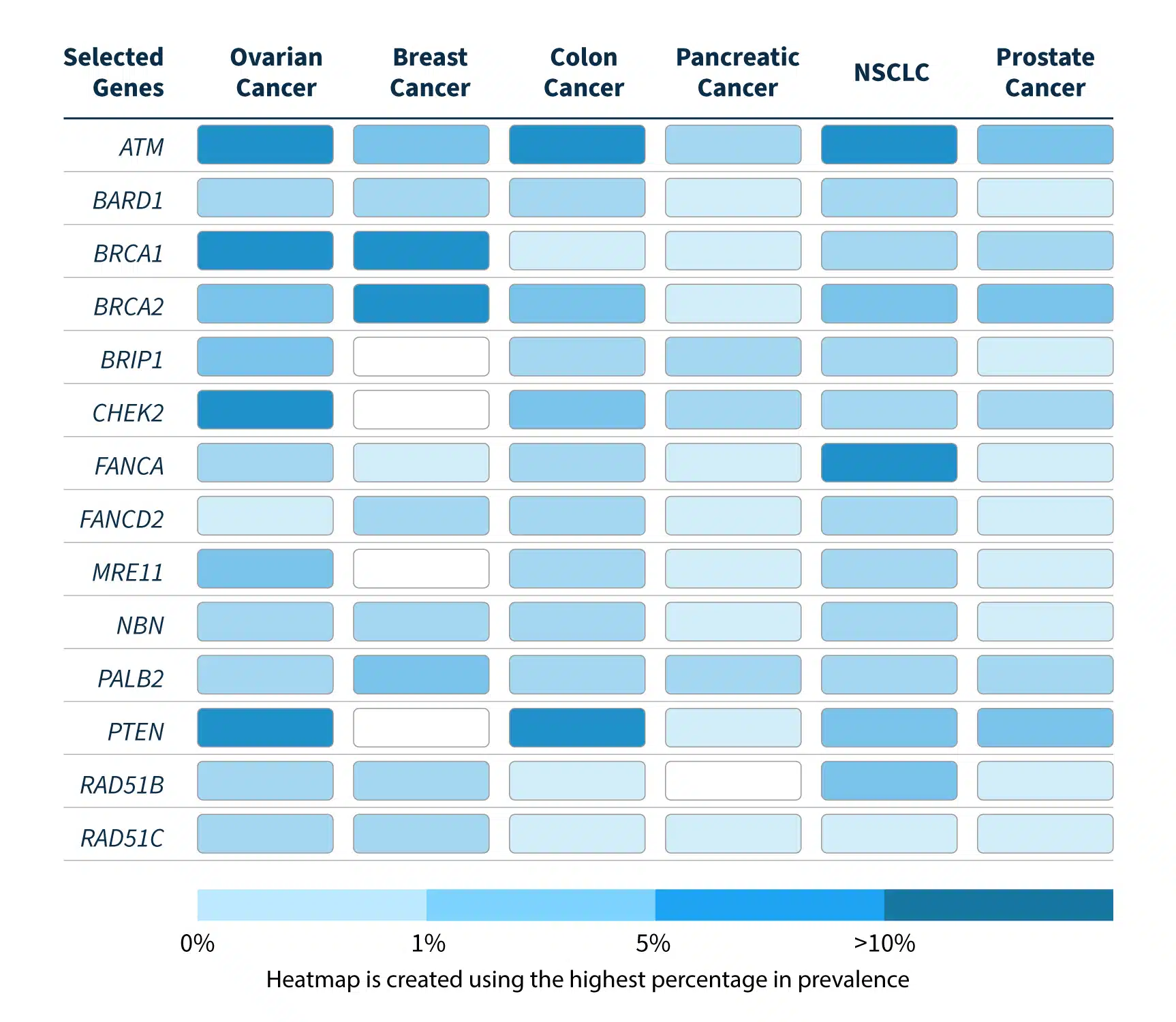
HRR gene mutations are prevalent in ovarian, breast, colon, pancreatic, lung, and prostate cancers4.
Comprehensive coverage of HRR genes
SOPHiA GENETICS offers three different HRR applications that enable accurate analysis of HRR status, covering up to 28 genes involved in the HRR pathway and encompassing SNVs, Indels, as well as gene amplifications.
SOPHiA DDM™ Homologous Recombination Applications are sample-to-report NGS-based applications that combine an expertly designed capture-based target enrichment kit with access to the analytical capabilities and interpretation-support functionalities of the SOPHiA DDM™ Platform.
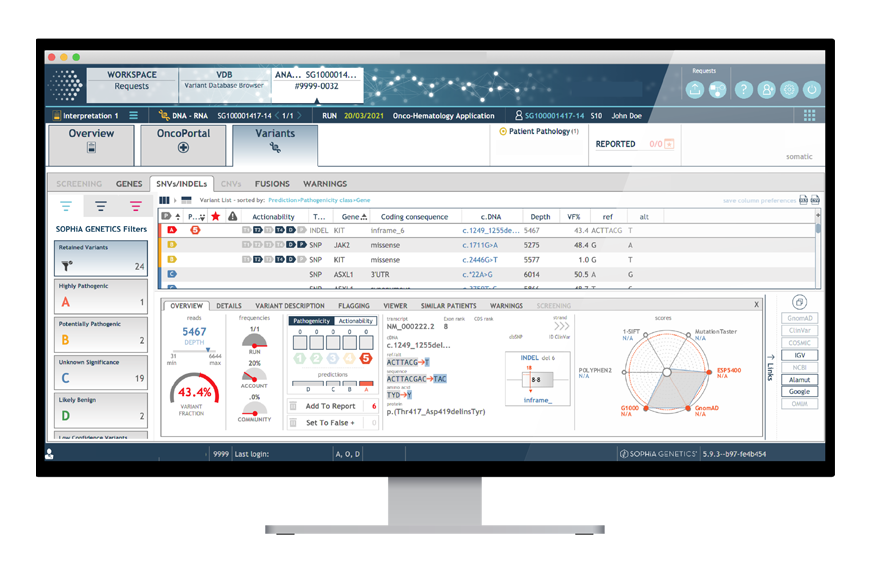
Intuitive features accelerating variant assessment
SOPHiA DDM™ Homologous Recombination Applications empower in-house expertise by providing high analytical performance and streamlined bioinformatics workflows with several intuitive features that accelerate variant assessment and interpretation.
The user-friendly interface provides accelerated filtering through:
- variant pathogenicity levels assigned using machine learning complemented by guideline-driven rankings.
- filtering by focusing on cancer type-specific variants.
- creating your own custom filtering strategies.
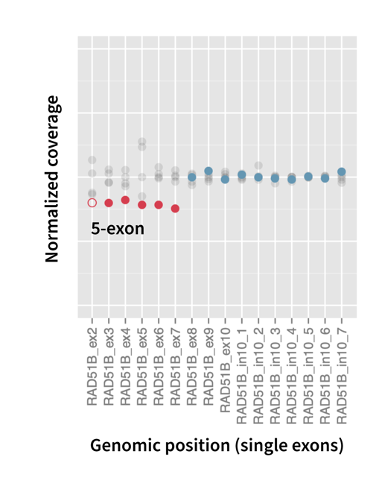
Detect gene amplifications confidently with our proprietary algorithm
Gene amplifications play a crucial role in HRD, as mutations in HRR genes lead to more error-prone repair throughout the genome1.
In addition, screening gene amplifications in FFPE samples is challenging because it is strongly affected by sample degradation and heterogeneity of the sample. To overcome this challenge, SOPHiA GENETICS has developed an algorithm designed to accurately identify gene amplifications, the breakpoint of which falls within a gene.
SNV: single nucleotide variations; Indel: Insertion and deletions.
Want to learn more about how to test for HRD in tumor samples
Discover our SOPHiA DDM™ HRD Solution and its deep-learning algorithm for detecting genomic scarring!
Specifications
| Parameters | SOPHiA DDM™ Mini Homologous Recombination Solution (HRS) | SOPHiA DDM™ HRS | SOPHiA DDM™ Extended HRS (Ext HRS) |
|---|---|---|---|
| Diseases Covered | Ovarian, prostate, breast, pancreas cancer | Ovarian, prostate, breast, pancreas cancer | Ovarian, prostate, breast, pancreas cancer |
| Target Region Size | 34 Kb | 66 Kb | 88 Kb |
| Key Biomarkers | 4 genes: BRCA1, BRCA2, RAD51B, TP53 | 16 genes: ATM, BARD1, BRCA1, BRCA2, BRIP1, CDK12, CHEK1, CHEK2, FANCL, PALB2, PPP2R2A, RAD51B, RAD51C, RAD51D, RAD54L, TP53 | 28 genes: AKT1*, ATM, BARD1, BRCA1, BRCA2, BRIP1, CCNE1, CDK12, CHEK1, CHEK2, ESR1*, FANCA, FANCD2, FANCL, FGFR1*, FGFR2*, FGFR3*, MRE11, NBN, PALB2, PIK3CA*, PPP2R2A, PTEN, RAD51B, RAD51C, RAD51D, RAD54L, TP53. |
| Sample Type | FFPE, fresh-frozen tissue | FFPE, fresh-frozen tissue | FFPE, fresh frozen tissue |
| Input Amount | 50 ng | 50 ng | 50 ng |
| Sequencer Compatibility |
|
|
|
| Library Preparation Time | 1.5 days | 1.5 days | 1.5 days |
| Analysis Time From FASTQ | From 4 hours | 4 hours | 4 hours |
| Detected Variants |
|
|
|
Resources
Want to know more?
Get in touch with us.
Our client services team is on hand to help.
References
1. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8914493/pdf/oyab053.pdf
- Pennington et al. Clin Cancer Res. 2014;20(3):764-75;
- The Cancer Genome Atlas Research Network. Nature. 2011;474(7353):609–15;
- Risch et al. J Natl Cancer Inst. 2006;98(23):1694-706.
- Mekonnen N, Yang H and Shin YK. Front Oncol. 202212:880643.