SOPHiA DDM™ for
SARS-CoV-2
Accelerate Covid-19 research and surveillance
The SOPHiA GENETICS SARS-CoV-2 analytical solutions enable rapid and precise identification of circulating and emerging variants, including Omicron.
Leveraging our extensive experience and knowledge in genomics, the SOPHiA DDM™ platform offers analysis of the whole viral genome for sensitive variant detection through streamlined, end-to-end bioinformatics workflows (including as early as sampling to surveillance reporting). These solutions accelerate surveillance and evolution studies of SARS-CoV-2.
SARS-CoV-2 solutions offer future-proof strain identification including*:
‘Omicron’ (B.1.1.529), ‘Alpha’ (B.1.1.7), ‘Beta’ (B.1.351), ‘Gamma’ (P.1), ‘Epsilon’ (B.1.429), ‘Iota’ (B.1.526), ‘Kappa’ (B.1.617), ‘Mu’ (B.1.621)
Featuring Omicron variant detection: optimized identification and filtering for current and emerging variants
Product Details
Identify new and emerging variants with confidence
SOPHiA GENETICS SARS-CoV-2 solutions are fast, highly sensitive and analytically accurate to surveil circulating, emerging variants on a global scale. The overall clade distribution in the SOPHiA DDM™ platform is similar to results from global datasets which require manual analysis and interpretation. Using the SOPHiA DDM™ platform, data quality is controlled, analyses can be automated, and data is screened in real time.
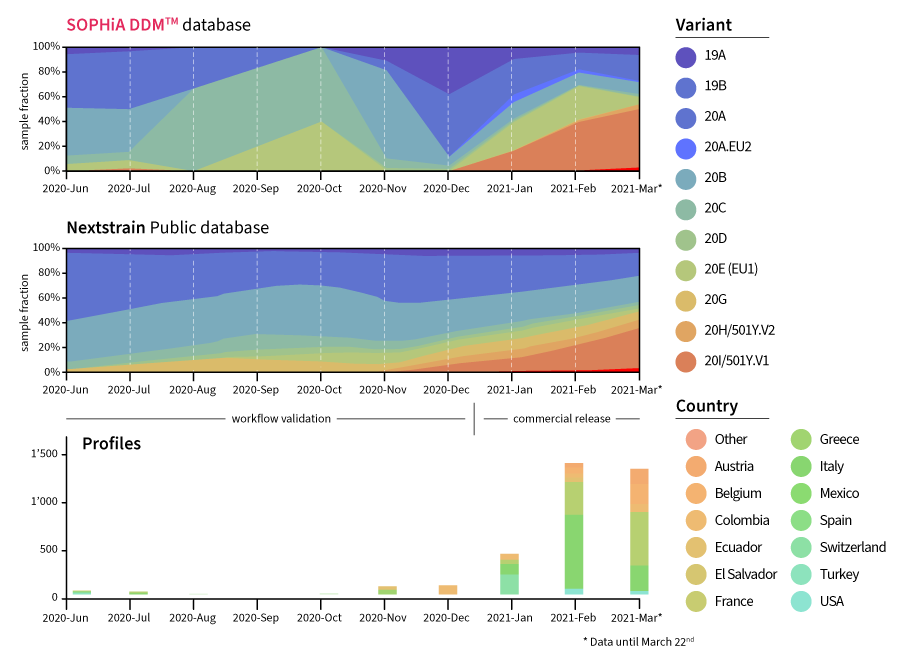
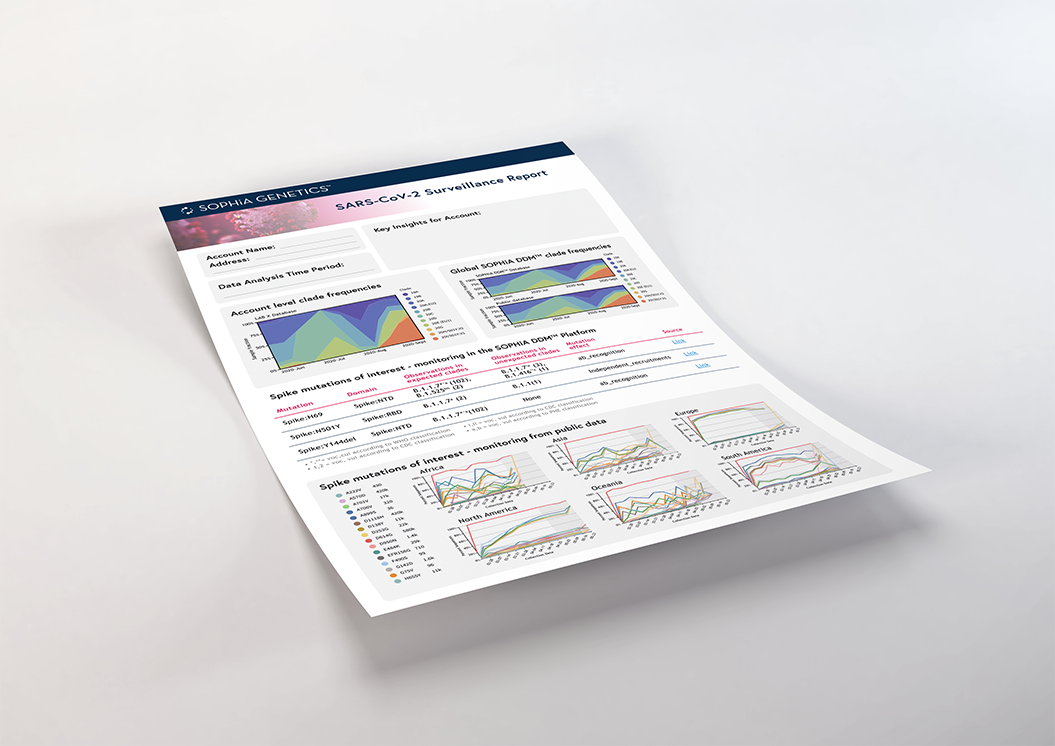
Real-time reporting of SARS-CoV-2 data with the SOPHiA GENETICS SARS-CoV-2 Surveillance Report
The SOPHiA DDM™ platform provides at-a-glance reporting to streamline the interpretation process for variants of concern (VOC), including valuable insights such as:
- Lineages associated with spike mutations
- Variant classification (VOC, VOI/VUI, VOHC, Monitoring) consistent with WHO/CDC and Public Health England
- Real-time tracking of known circulating variants by geography
- Hotspot identification by variant
- Key spike protein mutational monitoring
- New mutation and new mutation combination spotlights
- Insights enhanced with published scholarly data
Available SARS-Cov-2 Panels
| Feature / Application | Paragon Genomics Cleanplex® | Paragon Genomics Flex® | QIAseq® SARS-CoV-2 |
ARTICv3® SARS-CoV-2 |
ATOPlex® SARS-CoV-2 V1 |
|---|---|---|---|---|---|
| SARS-CoV-2 Detection | ✓ | ✓ | ✓ | ✓ | ✓ |
| Surveillance and Monitoring | ✓ | ✓ | ✓ | ✓ | ✓ |
| Variant Typing | ✓ | ✓ | ✓ | ✓ | ✓ |
| New Variant Research | ✓ | ✓ | ✓ | ✓ | ✓ |
| Library Preparation Kits Available | ✓ | ✓ | – | – | – |
Resources
Flyers
Find out more
Read more
- A straightforward molecular strategy to retrospectively investigate the spread of SARS-CoV-2 VOC202012/01 B.1.1.7 variant
- First reported cases of SARS-CoV-2 sub-lineage B.1.617.2 in Brazil: an outbreak in a ship and alert for spread
- SOPHiA Genetics and Paragon Genomics join forces
- Covid: ‘The virus is always searching for its next move’
Contact us
Please fill out the form below to get in touch