Menu
Menu
Explore this infographic summary to gain insights into the key findings from Pozzorini et al.’s publication on the GIIngerTM deep learning algorithm for prediction of HRD status and patient response to PARPi treatment in ovarian cancer.
Click here to read the full publication.
Pozzorini C, Andre G, Coletta C, et al. GIInger predicts homologous recombination deficiency and patient response to PARPi treatment from shallow genomic profiles. Cell Rep Med. 2023 Dec 19;4(12):101344. doi: 10.1016/j.xcrm.2023.101344.
GIInger™ data were generated using the SOPHiA DDM™ Dx HRD Solution, available as a CE-IVD product for In Vitro Diagnostic Use in European Economic Area (EEA), the United Kingdom and Switzerland . SOPHiA GENETICS products are for Research Use Only and not for use in diagnostic procedures unless specified otherwise.
Do you want to further your understanding of overfitting, or are you interested to learn how we avoid overfitting when developing predictive machine learning models for clinical research? Browse our tech note, which includes a step-by-step, published example of how we developed a model that was able to successfully support the evaluation of kidney cancer tumor upstaging in individual patients.
Genomic instability is a hallmark of cancer and targeting its mechanisms has helped inform effective therapeutic strategies1,2. However, there are limitations with current methods of genomic instability assessment. Here, we explore genomic instability in the context of homologous recombination deficiency and the value of deep learning-based methods of detection.
The DNA in our cells endure up to one million damaging events each day, caused by both exogenous (i.e. environmental) and endogenous (i.e. internal metabolic) factors3. These events activate a complex network of DNA damage response (DDR) pathways, which facilitate DNA repair and maintain the stability of the genome4. Mutations and/or dysfunction in DDR pathways can lead to unrepaired DNA damage, resulting in genomic instability4. Genomic instability, in turn, increases the cell’s propensity for genetic alterations that cause cancer initiation and progression4,5.
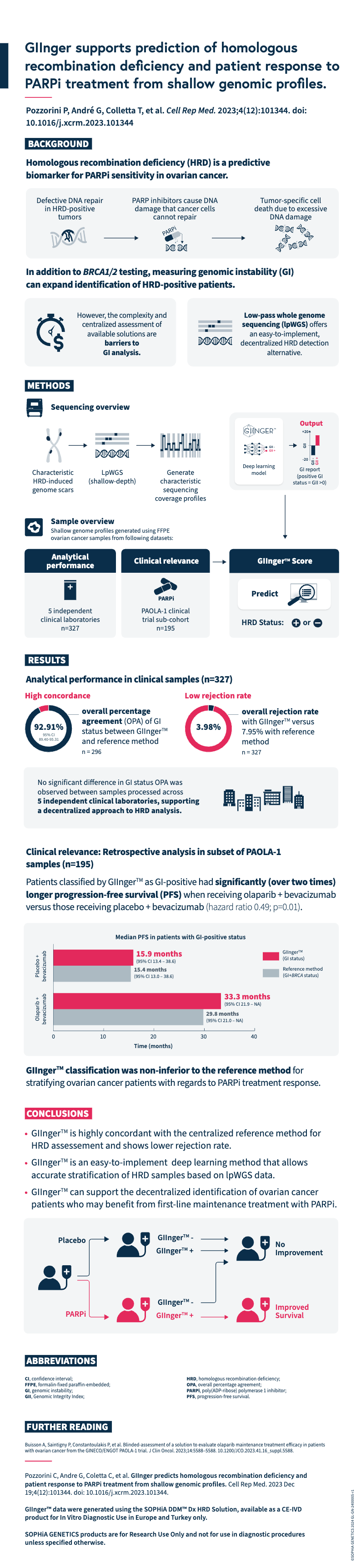
One of the major DDR pathways is the homologous recombination repair (HRR) pathway, responsible for repairing double-strand breaks (DSBs) in DNA5. Loss of function in HRR, known as homologous recombination deficiency (HRD), causes cells to rely on error-prone DNA repair pathways, resulting in the accumulation of genetic aberrations that lead to genomic instability5 (Fig 1). HRD is a well-established prognostic and predictive biomarker in different cancer types (e.g. ovarian, breast, prostate, and pancreatic)5–7.
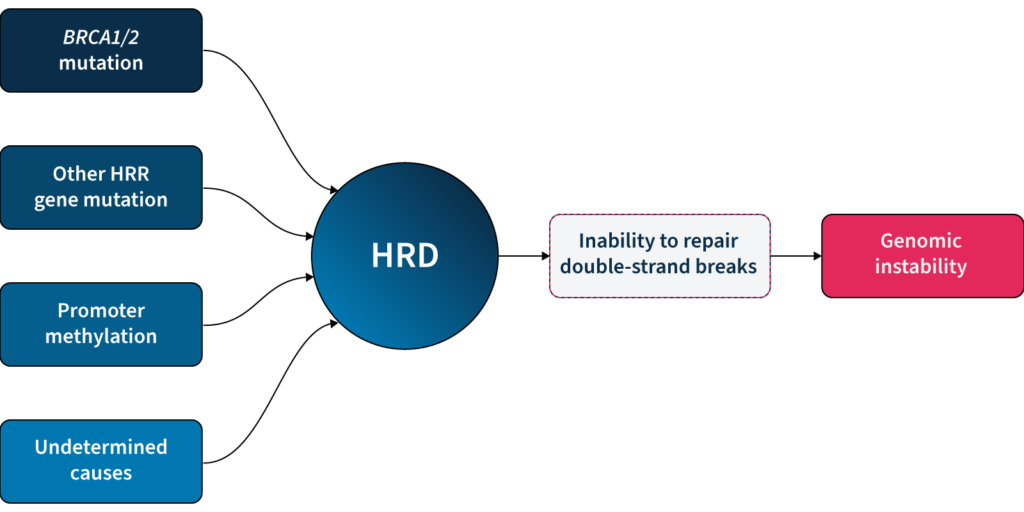
HRD-positive tumors are sensitive to targeted inhibition of poly-ADP ribose polymerase (PARP), key proteins involved in DSB repair7. By blocking PARP, the HRD-positive cell can no longer rely on error-prone pathways for DSB repair and the cell dies, a process known as ‘synthetic lethality’5,6. PARPi therapy has revolutionized the management of HRD-positive patients with advanced ovarian cancer, significantly improving progression-free survival when used as a first-line maintenance therapy8. PARPi therapies also have approved indications in breast, pancreatic and prostate cancer9, with trials underway in other cancer types, such as colorectal10.
Based on the predictive value of HRD status for PARPi benefit, clinical guidelines recommend HRD testing in patients with advanced ovarian cancer7,11,15. HRD status can be determined by examining 1) the underlying causes of HRD, and 2) the effect of HRD, i.e. genomic instability5,7. The most well-known causes of HRD are loss-of-function mutations in HRR genes, including BRCA1 and BRCA25,7. However, loss-of-function in HRR genes is diverse amongst patients12, making patient stratification solely based on genotyping challenging. Also, approximately 30–40% of HRD cases are due to unknown causes13,14. Measuring genomic instability allows the assessment of HRD, regardless of its underlying etiology5,7.
Genomic instability status can help identify a sub-group of women who are BRCA wild-type but may still derive benefit from PARPi therapy15. By measuring genomic instability, clinicians and researchers can therefore go beyond HRR mutation detection and expand the potential benefit of PARPi in patients.
Many methods for measuring genomic instability rely on the identification of specific mutational signatures or genomic ‘scars’ associated with large-scale structural rearrangements in chromosomes. In HRD-positive cancers, the characteristic genomic scars are loss of heterozygosity (LOH), large-scale state transitions (LST), and telomeric-allelic imbalance (TAI)16–18.
Click the boxes below to learn more:

A cross-chromosomal event that results in loss of part of a gene or entire gene(s) and the surrounding chromosomal region.

Chromosomal breaks between adjacent regions of at least 10 Mb.
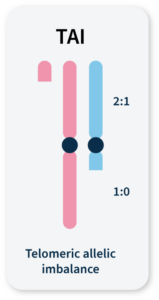
Accumulation of a discrepancy in the 1:1 allele ratio at the end of the chromosome (telomere).
The combined number of LOH, LST, and TAI events generate a genomic instability score (GIS) that reflects the level of genomic instability. Some commercially available HRD tests combine tumor BRCA mutation testing with a GIS5,19. Methods that integrate multiple genome-wide signatures (e.g. HRDetect) are among the most promising for detecting HRD status7,20. However, both GIS and HRDetect methods require deep genomic profiling data (>30x coverage), which can be costly and difficult to implement in routine analysis.
Alternative approaches that rely on the detection of copy number changes from WGS at low (~1x) sequencing depth (low-pass WGS) can predict tumor HRD status21,22 and provide an affordable and easy-to-implement HRD detection method. However, the sensitivity of existing methods that solely rely on this type of genomic scar to identify HRD samples is limited, and their utility in a clinical context remains untested22.
Unlocking the full potential of low-pass WGS in HRD detection requires going beyond the enumeration of biomarker events and examining alternative features of the cell that can result from genomic instability.
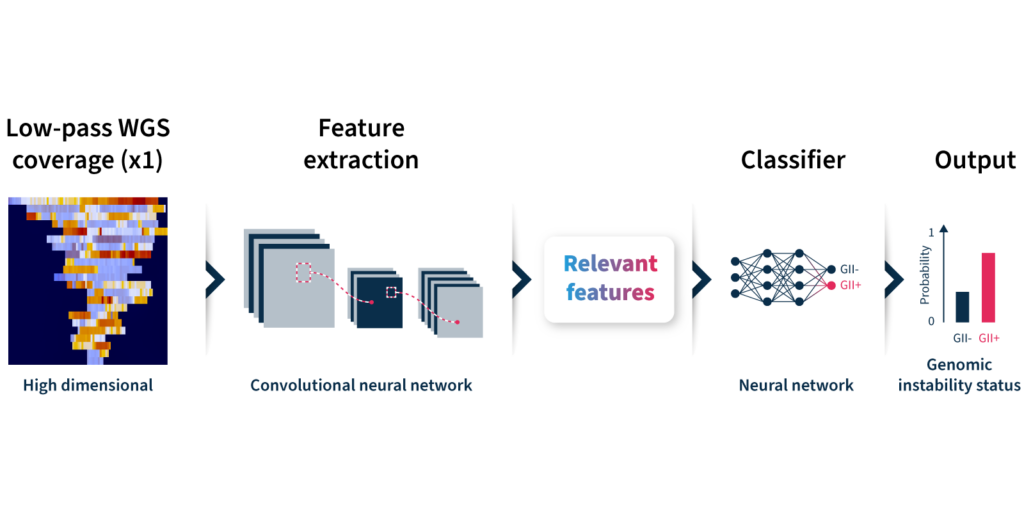
To overcome the limitations of current genomic instability measures, our expert team at SOPHiA GENETICS developed the GIInger™ algorithm exclusively available on the SOPHiA DDM™ Platform. GIInger™ is a deep learning-based approach to measuring genomic instability in ovarian cancer samples. Rather than relying on the enumeration of biomarkers events, GIInger™ leverages differences in the spatial distribution of genomic scars in low-pass WGS coverage profiles23.
Let’s take a closer look at how the algorithm predicts genomic instability status (Fig 2):
Unfamiliar with deep learning terminology? Read our guide on machine learning jargon.
By adopting GIInger™ into next generation sequencing (NGS) workflows, clinical researchers can benefit from an in-house, affordable approach to genomic instability measurement. The SOPHiA DDM™ Platform offers applications that enable laboratories to easily implement GIInger™ into their routine NGS analysis:
Want to see how GIInger™ can help maximize insights from your data? Get in touch with our team and request a demo.
The term « SOPHIA » used by the speaker refers to SOPHiA GENETICS and its products. The opinions expressed during this presentation are these of the speaker and may not represent the opinions of SOPHiA GENETICS. SOPHiA GENETICS does not provide support in the validation of custom products for clinical use. SOPHiA DDM™ Dx Homologous Recombination Deficiency Solution is available as a CE-IVD product for In Vitro Diagnostic Use in the European Economic Area (EEA), the United Kingdom and Switzerland. SOPHiA GENETICS products are for Research Use Only and not for use in diagnostic procedures unless specified otherwise.
SOPHiA DDM™️ Dx ROS is a novel CE-IVD marked NGS assay for detecting gene fusion and exon skipping events from small lung cancer biopsy samples. A performance evaluation study demonstrates that, compared with existing RNA-based assays, SOPHiA DDM™️ Dx ROS provides accurate actionable clinical insights for healthcare professionals to make disease management decisions for patients with advanced lung cancer
At ESMO 2022, oncology experts gathered in Paris and online to share and debate the new developments in the field of medical oncology. This year's program featured more than 20 tracks covering all tumor types, therapeutic innovations, translational research, patient advocacy, public policy, and more...
Discover our summary of three compelling talks showcasing the journey towards precision therapy in various tumor types.
Genomic profiling and molecular targeting of lung cancer brain metastases1
Haiying Cheng, Dept. Medical Oncology, Montefiore Medical Center/Albert Einstein College of Medicine, Bronx, NY, USA
Approximately 57% of patients with non–small-cell lung cancer (NSCLC) present with metastatic disease2. Among them, brain metastases (BM) affect up to 45% of all cancer patients and arise from lung cancers in 40-50% of the cases3. There have been limited studies investigating the genetic signatures of LC BM, and with small cohorts so far.
Assembling a large number of lung cancer cases (47215 NSCLC; 29438 lung adenocarcinoma), Dr Cheng and colleagues looked for key genetic alterations in loco-regional lesions (Loco), extracranial metastases (EM), and BM with comprehensive genomic profiling (CGP). They found significantly more genetic alterations in the PI3K/AKT/mTOR pathway in BM (Loco 13.0% vs EM 14.5% vs BM 18.1%), primarily driven by RICTOR amplification (Loco 3.6% vs EM 6.2% vs BM 8.6%).
RICTOR amplification is the most enriched actionable genomic target in NSCLC brain metastases.
Furthermore, in vitro genetic knockdown and pharmacological inhibition of RICTOR significantly reduced migration and invasion in RICTOR-amplified NSCLC cells, whereas RICTOR upregulation promoted these processes, modulating the AKT, MET, EMT, and CXCL12 chemokine-CXCR4 pathways. Finally, in vivo studies in orthotopic mouse models revealed that both RICTOR and mTOR1/2 inhibition significantly reduced lung cancer tumor growth and spread in the brain.
Dr Cheng provided evidence for the benefit of further investigation on the development of RICTOR-targeted therapeutic strategies for the treatment and/or prevention of lung cancer BM. This study is a good example of how genomic profiling, combined with functional analyses, can identify new potential therapeutic targets.
Neoadjuvant immune checkpoint inhibition in locally advanced MMR-deficient colon cancer: The NICHE-2 study4.
Myriam Chalabi, Gastrointestinal Oncology, Netherlands Cancer Institute, Amsterdam, Netherlands
Mismatch repair deficiency (dMMR) is observed in ~15% of colorectal cancers (CC)5 and 1/3 is associated with Lynch Syndrome. This characteristic genetic signature is marked by high levels of microsatellite instability (MSI) and resistance to standard-of-care neoadjuvant chemotherapy (5-7% pathological response (≤50% residual viable tumor; PR)). NICHE-1 exploratory study showed the potential of neoadjuvant immunotherapy in patients with dMMR CC with extraordinary PR in 100% of the patients6.
Dr Chalabi presented the NICHE-2 investigator-initiated study, conducted in 6 hospitals in the Netherlands. 107 patients with non-metastatic untreated dMMR CC and mainly high-risk tumors received injections of nivolumab and ipilimumab within 6 weeks prior surgery. The impressive pathological tumor regression was shown in a waterfall plot that led to a standing ovation! 95% of the treated patients showed PR, and 67% had no residual viable tumor (complete PR; cPR), contrasting with the neoadjuvant chemotherapies in the same patient population. Only 4% experienced grade 3-4 immune-related adverse events and 98% of patients underwent timely surgery, meeting the safety primary endpoint. To date, no disease recurrence has been observed and the 3 years disease-free survival data are expected next year.
Neoadjuvant immunotherapy has the potential to become standard of care for patients with dMMR colon cancer.
NICHE-2 trial opens the possibility that a surveillance approach may be possible for some patients with early dMMR CC and gives a glimpse at the potential of translational research to identify predictive biomarkers in pre- and post-treatment samples. While those preliminary results are extremely promising and we surely wait for the longer-term follow-up data to confirm them, patient selection remains crucial. Indeed, neoadjuvant decisions are based on radiological assessment of the tumor, particularly difficult in dMMR cancers, as well as the biopsy, and Dr Chalabi highlighted the need for improvement in the imaging techniques and circulating DNA analyses.
Final overall survival results from the phase III PAOLA-1/ENGOT-ov25 trial evaluating maintenance olaparib plus bevacizumab in patients with newly diagnosed advanced ovarian cancer7.
Isabelle Ray-Coquard, Department Of Medical Oncology, Centre Léon Bérard, and GINECO, Lyon, France
The late diagnosis of advanced ovarian cancer (AOC) is often accompanied by relapse, despite surgery and platinum-based chemotherapy. Treatment with olaparib (ola), a poly(adenosine diphosphate–ribose) polymerase inhibitor (PARPi), provided progression-free survival (PFS) benefit as maintenance therapy in patients with AOC carrying mutations in BRCA1 or BRCA2 (BRCAm)8. Besides, the incorporation of the antiangiogenic agent bevacizumab (bev) is a recognized option in addition to chemotherapies9.
PAOLA-1 investigators conducted a phase III trial where 806 patients with AOC and after first-line platinum-based chemotherapy plus bev were randomly assigned in a 2:1 ratio to ola + bev or placebo (pbo) + bev treatment. The primary endpoint was the PFS. They showed that combined treatment with ola + bev reduced the risk of relapse by 41% compared to bev alone, reducing by 67% in HRD+ patients (exhibiting BRCAm and/or genomic instability score ≥42)10.
Here, Dr Ray-Coquard presented the final overall survival (OS) results, a key secondary endpoint. She showed that the OS rate after 5 years was not different between the two arms (47.3% vs 41.5%) but significantly increased for HRD+ patients treated with ola + bev (65.5% vs 48.4%), regardless of BRCAm status. Also, PFS was significantly increased in the same population (46.1% vs 19.2%).
Maintenance therapy with olaparib plus bevacizumab improved survival in HRD+ patients with newly diagnosed advanced ovarian cancer.
With the absence of new safety signals and major adverse effects, these data confirmed the benefit of olaparib and bevacizumab combination as a standard of care for HRD+ patients and reinforced the importance of precision medicine and biomarker testing to guide treatment decisions.
Meet the new SOPHiA DDM™ Dx RNAtarget Oncology Solution (ROS).
Cancer management outcomes can strongly benefit from robust and accurate detection of gene fusions. SOPHiA GENETICS developed the SOPHiA DDM™ Dx RNAtarget Oncology Solution (ROS) to make this possible.
Gene fusions are “hybrid genes” formed by the fusion of two genes into one and producing a novel protein. These are mostly due to DNA translocations, but also sometimes resulting from DNA insertions, inversions, duplications, or deletions1. Exon-skipping events, the erroneous excision of one or more exons during RNA splicing, also lead to pathologically structured genes. Both play an important role in carcinogenesis1 and, because of this, have been preferential targets of precision medicine as well as of some of the most effective treatments in fusion-positive cancers2,3.
Whether specific rearrangements occur at DNA or RNA level, they can both lead to new proteins with altered functions: in human cancers, more than 10,000 gene fusions creating oncogenic drivers of specific tumors or activators of oncogenic signalling have been identified4. Given the large number of gene fusions to identify, researchers worldwide increasingly need intelligent bioinformatics tools to automate the process - and even more: detect them with suitable technical benefits, like the possibility of customizing gene panels, the use of a minimum input sample, and the chance to optimize the lab workflow using high sensitivity technology. Considering the importance of gene fusions as a diagnostic and prognostic marker for cancer management in the clinical setting5-7, these should be transitioned from the research to patient's bedside. In this particular context, the precious RNA sample for gene fusion testing is not only a “simple sample” - it represents a unique and individual opportunity for better treatment outcomes8. The right technology choice sets the tone for high-accurate detection of gene fusions and, consequently, for high-precision medicine.
Moreover, the possible association between gene fusion type and tumor phenotype can flag the clinical value of accurate gene fusion detection for:
The right technology choice sets the tone for high-accurate detection of gene fusions and, consequently, for high-precision medicine.
Multiple approaches have been developed to effectively detect gene fusions at DNA, RNA, and protein level based on their significant clinical relevance. The investigation of gene fusions at the DNA level allows to identify structural chromosomal rearrangements, while looking at RNA uncovers fusions generated during RNA splicing without any chromosomal rearrangement and potentially translated into defective fusion proteins. Gene fusion identification has been traditionally performed by immunohistochemistry, fluorescence in situ hybridization (FISH), or polymerase chain reaction (PCR), but next-generation sequencing (NGS) methods represent the current gold standard6,7. These high-throughput sequencing technologies can identify gene fusions either at DNA or RNA level. NGS has become the technology of choice for fusion detection in solid tumors in agreement with international guidelines7. The simultaneous analysis of detailed (nucleotide-level) and comprehensive (genome-wide) information on the genome or transcriptome (all RNA transcripts) allow searching for the constantly increasing number of fusion genes, which would be otherwise impossible to achieve without high-throughput techniques8,9.
Although DNA-based sequencing technologies perform a comprehensive investigation, their short-read NGS approach currently hinders the proper mapping of several gene fusions. Furthermore, they require a considerable amount of sequencing, storage space, and long computational analysis, which makes them expensive and time-consuming. On the other hand, RNA-based approaches are more cost-effective: less storage space and analysis time are needed since only genetic regions which are transcribed and spliced into mature mRNA are explored; therefore, expressed gene fusions are exclusively detected. Also, RNA-based approaches overcome possible technical limitations of DNA-based methods derived from the localization of fusion breakpoints either within long DNA intronic regions (difficult to capture) or on different sites on DNA and RNA encoding the same fusion because of post-transcriptional and splicing rearrangements.
The success of RNA-based gene fusion detection approaches relies on sample quality and quantity: proper RNA extraction, stability, abundance of mRNA transcribed from the gene of interest, and a sufficient amount of sample can often be challenging9. Technics using small inputs coming from precious biopsy samples are required. Furthermore, algorithms with high gene fusion prediction accuracy are crucial. Finally, the complexity of the analysis, which is computationally demanding, remains a significant challenge10.
SOPHiA GENETICS aimed to solve the current challenges of RNA-based approaches for gene fusion detection by developing the SOPHiA DDM™ Dx RNAtarget Oncology Solution (ROS).
Here we list seven reasons11 why you should consider SOPHiA DDM™ Dx ROS for your assays:
Compared to a fully-guided amplicon-based approaches, the capture-based design of SOPHiA DDM™ Dx ROS detects fusion without prior knowledge of the partners, increasing considerably the power of fusion detection and exon-skipping events.
SOPHiA DDM™ Dx ROS requires only 10 ng of RNA from formalin-fixed paraffin-embedded (FFPE) or fresh frozen tissues. No fragmentation is required before FFPE samples processing, while a short simple fragmentation is performed on fresh frozen tissues. The solution is also compatible with total nucleic acid (TNA).
Together with gene fusions, SOPHiA DDM™ Dx ROS can detect single nucleotide variants (SNVs) and insertion/deletion mutations (Indels). The SNV/Indels calling performed on RNA indicates expression and, therefore, the related biological relevance of the detected mutations is also provided.
The analytical requirements of SOPHiA DDM™ Dx ROS have been adapted to balance the solution's sensitivity and specificity. SOPHiA DDM™ Dx ROS variant caller identifies PCR duplicates to enhance results accuracy. The analytical requirements were optimized to balance the sensitivity and specificity of our technology. The analytical parameters were carefully chosen to balance the need to remove deamination artifacts and background noise. Furthermore, SNV calling algorithms account for biases due to the low biological expression level of the gene of interest. To achieve global standard results with high sensitivity detection of gene fusions, SOPHiA DDM™ Dx ROS was designed based on the recent ESMO (Tier I and II) and NCCN international guidelines for lung cancer6. The probabilistic model applied to the data leverages multiple features to deliver results with reduced false positives and high accuracy of fusion calling.
SOPHiA DDM™ Dx ROS is easily adaptable to the unique needs of your laboratory. An optimized workflow with a dedicated probe design process and extensive wet lab QC experiments provides you with a ready-to-use panel, reducing the need for additional optimization in the lab.
SOPHiA DDM™ Dx ROS comes with a streamlined 1.5-day workflow that supports the whole analysis and ensures access to record-time results.
The SOPHiA DDM™ Platform fully supports you in the data analysis and interpretation process: pipelines are adapted to sample type, sequencer, enrichment kit, and chemistry used in your laboratory. Moreover, s streamlined workflow helps you to annotate, interpret, and report relevant NGS variants. The complexity of the analysis is minimized. The intuitive and user-friendly interface as well as its accelerated data visualization and interpretation and its customizable reporting and comprehensive QA report increase the efficiency of the decision-making process. The OncoPortal™ Plus module, gathering the latest scientific evidence and guidelines on the actionability and significance of each genomic alteration, completes the all-around support of the SOPHiA DDM™ Platform to facilitate your interpretation process.
SOPHiA DDM™ Dx ROS supports your laboratory in gene fusion detection. With its high sensitivity and accuracy, it allows the identification and the expression assessment of novel gene fusions, SNVs and Indels. It can also adapt to your research needs thanks to a fully customizable gene panel. Moreover, a streamlined workflow and the SOPHiA DDM™ Platform support you throughout the whole assay, from the RNA sequencing to the data analysis and interpretation. With SOPHiA DDM™ Dx ROS, you will maximize the insights from precious biopsy specimens getting highly reliable results even from samples of limited quantity.
1. Mitelman F, Johansson B, Mertens F (eds.) (2016) Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer,.http://cgap.nci.nih.gov/Chromosomes/Mitelman.
2. Taniue K, Akimitsu N. Fusion Genes and RNAs in Cancer Development. Noncoding RNA. 2021 Feb 4;7(1):10.
3. Brien GL, Stegmaier K, Armstrong SA. Targeting chromatin complexes in fusion protein-driven malignancies. Nat Rev Cancer. 2019 May;19(5):255-269.
4. Bruno R, Fontanini G. Next Generation Sequencing for Gene Fusion Analysis in Lung Cancer: A Literature Review. Diagnostics (Basel). 2020 Jul 27;10(8):521.
5. Mertens F, Johansson B, Fioretos T, Mitelman F. The emerging complexity of gene fusions in cancer. Nat Rev Cancer. 2015 Jun;15(6):371-81.
6. National Comprehensive Cancer Network®. NCCN Clinical Practice Guidelines in Oncology. Non-Small cell Lung Cancer. Version 3.2022
7. Mosele F, et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol. 2020 Nov;31(11):1491-1505.
8. Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011 Feb;12(2):87-98.
9. Kaya C, et al. Limitations of Detecting Genetic Variants from the RNA Sequencing Data in Tissue and Fine-Needle Aspiration Samples. Thyroid. 2021 Apr;31(4):589-595.
10. Uhrig S, et al. Accurate and efficient detection of gene fusions from RNA sequencing data. Genome Res. 2021 Mar;31(3):448-460.
11. Data on file
At ASCO 2022, oncology professionals gathered in Chicago, Illinois, and online to discuss the latest advances in research and care for patients with cancer. This year's program featured over 200 sessions on Advancing Equitable Cancer Care Through Innovation. The presentations spanned from care delivery and regulatory policy to developmental therapeutics, gastrointestinal cancer, lung cancer, pediatric oncology, and beyond. Here, discover our summary of four outstanding ASCO presentations focusing on breast cancer treatment, diagnosis and follow-up, showcasing the power of precision medicine in healthcare.
Targetable genomic mutations in young women with advanced breast cancer1.
Norin Ansari, Yale New Haven Hospital, New Haven, CT
Advanced breast cancers (BC) in young women (under 40 years old) are often more aggressive and with worse prognoses than in older women. As treatment strategies can be dictated by the type of genomic alteration (GA), knowledge of BC genetic profiles across ages can greatly improve guidance and outcomes. In her poster presentation, Norin Ansari analyzed over 2,000 BC using hybrid-capture based comprehensive genomic profiling (CGP) to evaluate subtypes of GA and confirmed via immunohistochemistry (IHC) hormone receptors (HR) and PD-L1 status.
The study showed a mutations stratification within the population of BC depending on patient's age. Indeed, younger patients had higher rates of BRCA1, BRCA2, and RB1 mutations and lower rates of CDH1 and PIK3CA mutations than did older patients. Differences were statistically significant in BRCA1, CDH1, and PIK3CA. Norin Ansari also showed that breast tumors in younger women were less likely to be estrogen receptor positive (ER+) and more likely to be triple negative while no clear age-related pattern for HER2 status could be highlighted. Finally, younger women were more often PD-L1 positive and had lower tumor mutational burden (TMB) than their older counterparts.
Different mutational profiles may support differential use of targeted and immune therapies.
With increasing availability of targeted and immune therapies, knowing which GA each group of women has allows to better tailor therapies and leads to more effective treatments. For instance, BRCA1 mutations may lend to PARP inhibitor use while PIK3CA mutations may indicate the benefit of alpelisib prescription. The difference in genetic mutations between age groups can give a head start when treating women with breast cancer and CGP can refine the approach for better results.
Alpelisib + fulvestrant in patients with hormone receptor–positive, human epidermal growth factor receptor 2–negative advanced breast cancer: Biomarker analyses by next-generation sequencing from the SOLAR-1 study2.
Dejan Juric, Massachusetts General Hospital Cancer Center, Harvard Medical School, Boston, MA
PIK3CA mutations account for approximately 40% of the hormone receptor positive (HR+), HER2-negative (HER2-) advanced BC. PIK3CA encodes for a subunit of PI3K, key of a highly interconnected pathway regulating growth and cell survival. PI3K pathway alterations are associated with endocrine therapy resistance, hence the poor prognosis for HR+, HER2– advanced BC.
Dejan Juric introduced the SOLAR-1 phase 3 study, a randomized controlled study testing the efficacy of the combined administration of alpelisib (ALP, a PI3Kα-selective inhibitor and degrader) and fulvestrant (FUL, a selective estrogen receptor degrader) in HR+, HER2- advanced BC patients. SOLAR-1 shows improved progression-free survival (PFS) in ALP + FUL treated patients versus placebo + FUL of 11.0 and 5.7 months, respectively. Going one step further, they measured the efficacy outcomes in patients with specific gene alterations (GA) in a PIK3CA-altered cohort, applying a retrospective exploratory biomarker analysis.
SOLAR-1 baseline tumor samples were tested by next-generation sequencing (NGS) and clinical benefit was assessed using PFS and hazard ratio based on TMB and GA status in the PIK3CA-altered cohort. While ALP + FUL clinical benefit was seen across TMB quartiles, it was more pronounced in patients with a low TMB (PFS of 18.5 months with ALP versus 3.2 months with placebo). They also observed that, despite improved PFS with ALP + FUL treatment in all PIK3CA-altered patients, the level of benefit may depend on the mutation status of other genes involved in MAPK pathway, PI3K pathway, in endocrine therapy or CDK4/6 inhibitors resistance. For instance, greater benefit was observed with altered FGFR1/2 but limited in MYC- and RAD21-altered cohorts. Besides, ALP + FUL efficacy was independent of GA in TP53, ESR1, CCND1, MAP3K1 and ARID1A.
Clinical benefit of ALP + FUL was maintained regardless of alterations in most biomarkers.
To conclude, Dejan Juric showed that ALP + FUL treatment was beneficial in patients with HR+, HER2– advanced BC, especially with a low TMB, but that a comprehensive understanding of the unique mutational profile of each tumor via biomarkers analysis may explain the level of success and thus dictate further care.
Trastuzumab deruxtecan versus treatment of physician’s choice in patients with HER2-low unresectable and/or metastatic breast cancer: Results of DESTINY-Breast04, a randomized, phase 3 study3.
Shanu Modi, Memorial Sloan Kettering Cancer Center, Memorial Hospital, New York, NY, USA
Metastatic breast cancers (mBC) are classified according to the detection of certain receptors in the tumor cells, dictating the type of treatment to offer to the patients. Thus, mBC with an abnormally high quantity of human epidermal growth factor receptor-2 (HER2+) benefit from therapies targeting HER2 protein with monoclonal antibodies, while HER2- mBC receive treatment based on their HR status. However, the dichotomy between HER2+ and HER2- mBC does not suffice to find effective therapies for patients with low level of HER2 (HER2-low) currently treated as HER2-. The limited options and modest benefits of chemotherapy confirm the need for an adapted targeted strategy.
Trastuzumab deruxtecan (T-DXd) is part of a new generation of antibody-drug conjugates that delivers precision-focused chemotherapy directly to the cancer cells. Its activity was shown in tumors across a broad range of HER2 expression and a phase 1 trial showed promising efficacy of T-DXd in patients previously heavily treated with HER2-low mBC. Here, Shanu Modi presented us the DESTINY-Breast04 study, the first randomized phase 3 study of T-DXd for HER2-low mBC and its auspicious results.
Measuring the median progression-free survival (mPFS) in HR-positive mBC patients as primary endpoint, they observed statistically significant and clinically meaningful improvement for patients with HER2-low mBC compared to standard chemotherapy (mPFS 10.1 versus 5.4 months respectively: p<0.0001). Similar benefit was seen in all patients, regardless of their HR status, with T-DXd treatment through PFS and overall survival (OS) compared to standard chemotherapy.
DESTINY-Breast04 establishes HER2-low mBC as a targetable patient population with T-DXd as a new standard of care, with the potential to improve the survival for ~50% of all mBC patients.
We expanded the benefits of HER2 targeted therapy to a new population of breast cancer patients and established T-DXd as the new standard of care for HER2-low mBC.
These ground-breaking results presented at the 2022 ASCO Annual Meeting, and simultaneously published in the New England Journal of Medicine4, were acknowledged with a standing ovation from the audience of specialists. Anticipating these results to be practice changing, the study gives hope for many oncology professionals and patients.
Circulating tumor DNA and late recurrence in high-risk, hormone receptor–positive, HER2-negative breast cancer (CHiRP)5.
Marla Lipsyc-Sharf, Dana-Farber Cancer Institute, Boston, MA
Over half of metastatic recurrences in HR+ BC are late (occurring over 5 years from diagnosis) and thought to arise from minimal residual disease (MRD), a small number of cancer cells left in the body after treatment, hence the benefit of adjuvant therapy. MRD detection via circulating tumor DNA (ctDNA) is associated with high risk of BC recurrence in the early adjuvant setting across tumor subtypes. Little is known, however, about ctDNA for later settings.
Marla Lipsyc-Sharf presented the CHiRP prospective study of late recurrence in patients with high-risk HR+ BC without prior recurrence. 83 patients were followed with whole exome sequencing on primary tumor samples and plasma collection every 6-12 months to be processed with personalized RaDaRTM assay (12-51 variants) to detect ctDNA. Patients did not undergo routine surveillance body imaging or other circulating biomarkers testing. 68.7% of the patients had stage 3 disease and most received chemotherapy (90.4%) and adjuvant endocrine therapy (100%).
8 of 83 (10%) patients had detectable ctDNA at any timepoint during this study. As of last follow-up, 6 of them developed metastatic recurrence at various sites, 6-14 years after primary diagnosis, and one patient with detected ctDNA developed a locoregional recurrence.
All distant recurrences were detectable via ctDNA prior the recurrence with a median lead time of ~1 year.
Despite the low yet steady rate of recurrence in this small cohort with limited follow-up and infrequent plasma sampling (every 6-12 months), this study, published in Journal of Clinical Oncology6, shows that liquid biopsy can provide precious indication on the risk of relapse and thus point towards earlier intervention after MDR detection, improving patients' survival and quality of life.
1 https://meetings.asco.org/abstracts-presentations/210258
2 https://meetings.asco.org/abstracts-presentations/209230
3 https://meetings.asco.org/abstracts-presentations/209021
4 Modi S, Jacot W, Yamashita T, et al. Trastuzumab Deruxtecan in Previously Treated HER2-Low Advanced Breast Cancer [published online ahead of print, 2022 Jun 5]. N Engl J Med. 2022;10.1056/NEJMoa2203690. doi:10.1056/NEJMoa2203690
5 https://meetings.asco.org/abstracts-presentations/209216
6 Lipsyc-Sharf M, de Bruin EC, Santos K, et al. Circulating Tumor DNA and Late Recurrence in High-Risk Hormone Receptor-Positive, Human Epidermal Growth Factor Receptor 2-Negative Breast Cancer [published online ahead of print, 2022 Jun 4]. J Clin Oncol. 2022;JCO2200908. doi:10.1200/JCO.22.00908
Several RNA alterations have been described in Oncology, including gene fusions, recognized driver mutations in neoplasia1. More than 10,000 gene fusions have already been identified in human cancers1; it is estimated that up to 80% of solid tumors could benefit from gene fusion testing2. The number of new drugs specifically targeting gene fusion-positive cancers keeps growing: the advantages that proper gene fusion detection could bring to clinical cancer management are noticeable.
Transcriptome sequencing has emerged as an effective method to identify gene fusions and has become a routine task in cancer research and precision medicine3. However, although a variety of computational tools have been developed over the years, an optimal solution with high analytical performance for fusion detection and the ability to maximize the insights from small precious RNA samples has been lacking.
SOPHiA DDM™ RNAtarget Technology addresses these requirements by combining powerful novel (partner-agnostic) fusion detection capabilities as well as SNV/Indels detection in selected genes and expression changes assessment. Powered by a deep learning algorithm, the Technology works with a very low sample input, a fully customizable gene panel, and a streamlined automated workflow that supports all stages of the analysis with high sensitivity. Finally, a convenient yet powerful and fast results visualization and interpretation are ensured by the associated SOPHiA DDM™ Platform.
To better understand how SOPHiA GENETICS developed the SOPHiA DDM™ RNAtarget Technology and its features, we sat down with Mikhail Pertziger, the Clinical Application Product Manager for SOPHiA DDM™ RNAtarget Technology at SOPHiA GENETICS.
After studying Biomedical Science for my undergraduate degree, I continued with a PhD in the Molecular Biology of Breast and Colon Cancers, so Cancer and Molecular Diagnostics is very much the area where I have spent a lot of my academic and industry years. They say that the 21st century is the era of biology, and I will have to add that precision medicine is the future of cancer management. Biomarker-guided therapies are introducing dramatic differences in how oncology conditions are managed. Fusions are the latest frontier to receive broad applicability in the clinic with more and more drugs being introduced – this is bound to grow and accelerate. I'm excited to be working on a product that allows our customers to have access to a technology that takes on the learnings of the previous years and enables them to be more confident about detecting fusions.
I have worked at SOPHIA GENETICS for just over three years and currently lead the SOPHiA DDM™ RNAtarget Technology development and launch. The development team includes experts in a broad range of applications, including the core development of BioInformatics, NGS, programming, as well as logistics, Regulatory, Legal, and Marketing.
As I mentioned previously, gene fusions are the latest type of biomarker to receive broad applicability in cancer management. The results of targeting fusions reported in clinical trials and now being seen in routine care are fascinating. The number of new drug approvals in fusion-positive cancers has been continuously increasing over the last decade – up to 80% of solid tumors NGS tests could benefit from the inclusion of fusion testing. With histology-agnostic approvals, this number approaches 100%2. In parallel, more clinical trials are being rolled out to target fusion-positive cancers, hopefully leading to further improvements in treatment options in the near future.
Up to 80% of solid tumors NGS tests could benefit from the inclusion of fusion testing2
SOPHiA DDM™ RNAtarget Technology came about from our users' feedback on the need to have an application that allows them to detect novel fusions without sacrificing sensitivity in smaller biopsy samples. The work on this Technology started more than a year ago and has involved many feasibility and optimization studies to ensure that we're not just providing a regular solution, but a product that really helps users achieve more.
What we wanted to provide with this product was the ability for users to have a high-performance fusion detection that could be run in a very small amount of material, a streamlined (but robust) workflow, as well as the ability to not only detect fusions but also be able to extract as much information as possible from that small sample with detection of SNVs and Expression changes. For convenience, the gene content can be customized to fit the lab's individual needs, automated workflow to reduce resource constraints, and, finally, the product runs on the industry-leading SOPHiA DDM™ Platform, providing convenient visualization, annotation, and reporting of the results.
SNV detection in RNA is an intriguing area of development. There are several applications where SNV detection is beneficial, including the ability to run an RNA-only workflow in cases where genes of interest are sufficiently highly expressed. SNV detection in RNA also opens up the possibility of running RNA and DNA workflow sequentially, where the initial RNA workflow will likely detect the majority of relevant variants, leaving only a subset of samples that need to be also processed through the DNA workflow.
Another benefit of detecting SNVs in RNA is the ability to use them as an additional data point in calling SNVs in DNA or using RNA-based SNVs as a backup in case of issues with DNA.
Finally, having the information on SNV VF% in RNA, is like adding an additional dimension to the molecular profile created by DNA SNVs – as this provides more dynamic, rich, and potentially more insightful information on the state of things in the tumor.
Overall, there are many novel and unique ways this feature of SOPHiA DDM™ RNAtarget Technology could be used, and I'm excited to see how our users will utilize it.
One of the primary applications for the product is, of course, Lung cancer because of the limited amount of biopsy material that is generally available and a high number of clinically relevant fusions in this pathology. At the same time, the solution can be deployed to test any solid tumor, and we're looking at the possibility of running it in blood tumors as well. Moreover, because the gene content is entirely customizable, users can tailor the gene content to the needs of their labs, clinical research, or clinical trials that they want to be part of. This makes the application very versatile while removing the obstacle of manually optimizing the pipeline's performance because the SOPHiA GENETICS BioIT team will take care of that. Given the proliferation of tumor-agnostic biomarkers and, in particular, tumor agnostic fusions, the applicability of this Technology is only going to grow and expand in the future, and the fusion detection would become an integral part of genomic profiling of any cancer, together with SNV and CNVs.
SOPHiA DDM™ RNAtarget Technology can be deployed to test any solid tumor
It's really the combination of 3 main features that make for a cohesive and well-rounded product that offers a lot of value from several perspectives - Novel fusion detection, High sensitivity at low input amounts, and Customizability of the panel. These features provide an excellent foundation for a future-proof, high-performance solution. If you look at the market, there are very functional solutions that offer one or two of these features, but not all 3.
In addition to those three features, we included other functionality that I refer to as "two data points for every variant type," where 5’-3' imbalance serves as an additional data point for calling fusions, SNV detection in RNA can be used together with SNV calls in DNA, and expression changes provide further details and reassurance for the CNVs.
Finally, the streamlined protocol that can also be automated further refines the convenience factor of this solution.
"How" is straightforward: SOPHiA DDM™ RNAtarget Technology utilizes a hybrid-capture approach, which targets the key clinically relevant kinases. This protocol is augmented by a careful probe design process to make sure the panel performs at the highest level in the hands of our users.
On the other hand, it is worth highlighting that the detection of novel (or partner-agnostic) fusions is becoming a more and more prominent feature requested by labs striving to provide the highest level of care. This is underpinned by the higher inherent sensitivity, which is becoming more important in the rise in approval of fusion targeting therapies in a partner agnostic manner.
I would say it comes down to the challenges of needing to have high sensitivity in low sample input, the ability to detect novel fusions, and having a tailored solution that perfectly fits the needs of the lab, plus addressing the need for a convenient yet powerful visualization and interpretation platform – the SOPHiA DDM™ Platform.
SOPHiA GENETICS products are for Research Use Only and not for use in diagnostic procedures unless specified otherwise.
SOPHiA DDM™ Dx Hereditary Cancer Solution, SOPHiA DDM™ Dx RNAtarget Oncology Solution and SOPHiA DDM™ Dx Homologous Recombination Deficiency Solution are available as CE-IVD products for In Vitro Diagnostic Use in the European Economic Area (EEA), the United Kingdom and Switzerland. SOPHiA DDM™ Dx Myeloid Solution and SOPHiA DDM™ Dx Solid Tumor Solution are available as CE-IVD products for In Vitro Diagnostic Use in the EEA, the United Kingdom, Switzerland, and Israel. Information about products that may or may not be available in different countries and if applicable, may or may not have received approval or market clearance by a governmental regulatory body for different indications for use. Please contact us to obtain the appropriate product information for your country of residence.
All third-party trademarks listed by SOPHiA GENETICS remain the property of their respective owners. Unless specifically identified as such, SOPHiA GENETICS’ use of third-party trademarks does not indicate any relationship, sponsorship, or endorsement between SOPHiA GENETICS and the owners of these trademarks. Any references by SOPHiA GENETICS to third-party trademarks is to identify the corresponding third-party goods and/or services and shall be considered nominative fair use under the trademark law.