Menu
Menu

TNBC is a heterogeneous and aggressive subtype of breast cancer associated with poor outcomes. As a standard of care, patients with TNBC receive NAC therapy, allowing them to undergo breast-conserving surgery and achieve pCR, which is a strong predictor of favorable outcomes.
However, predicting pCR before treatment initiation remains a significant unmet need, showcasing the interest in having a method that can detect treatment response earlier in the patient journey. As an example, FDG-PET/CT has shown potential to detect residual disease early and predict poor outcomes in TNBC.
This proof-of-concept study, designed and developed by SOPHiA GENETICS in collaboration with AP-HP Hôpital Saint Louis and INSERM, aims to develop a multimodal machine learning (ML) model predictive of pCR, integrating multiple patient baseline parameters, such as clinical, histopathological, genomic, and PET imaging.
In this retrospective study, we have analyzed the data from 57 patients with stage II-III TNBC treated at Saint Louis Hospital, in Paris, between 2008 and 2015, with baseline PET and NAC followed by breast surgery.
A total of 241 baseline features were extracted from each patient, out of which 17 were selected for final analysis, encompassing 3 clinical, 2 histopathological, 3 molecular, 3 PET non-radiomic, and 6 radiomic parameters.
Using support vector (SVMs) with nested leave-pair-out cross-validation, the researchers developed an ML model optimized to predict pCR. Model performance was assessed using area under the curve (AUC), and Kaplan-Meier analysis was used to explore associations with event-free survival (EFS).
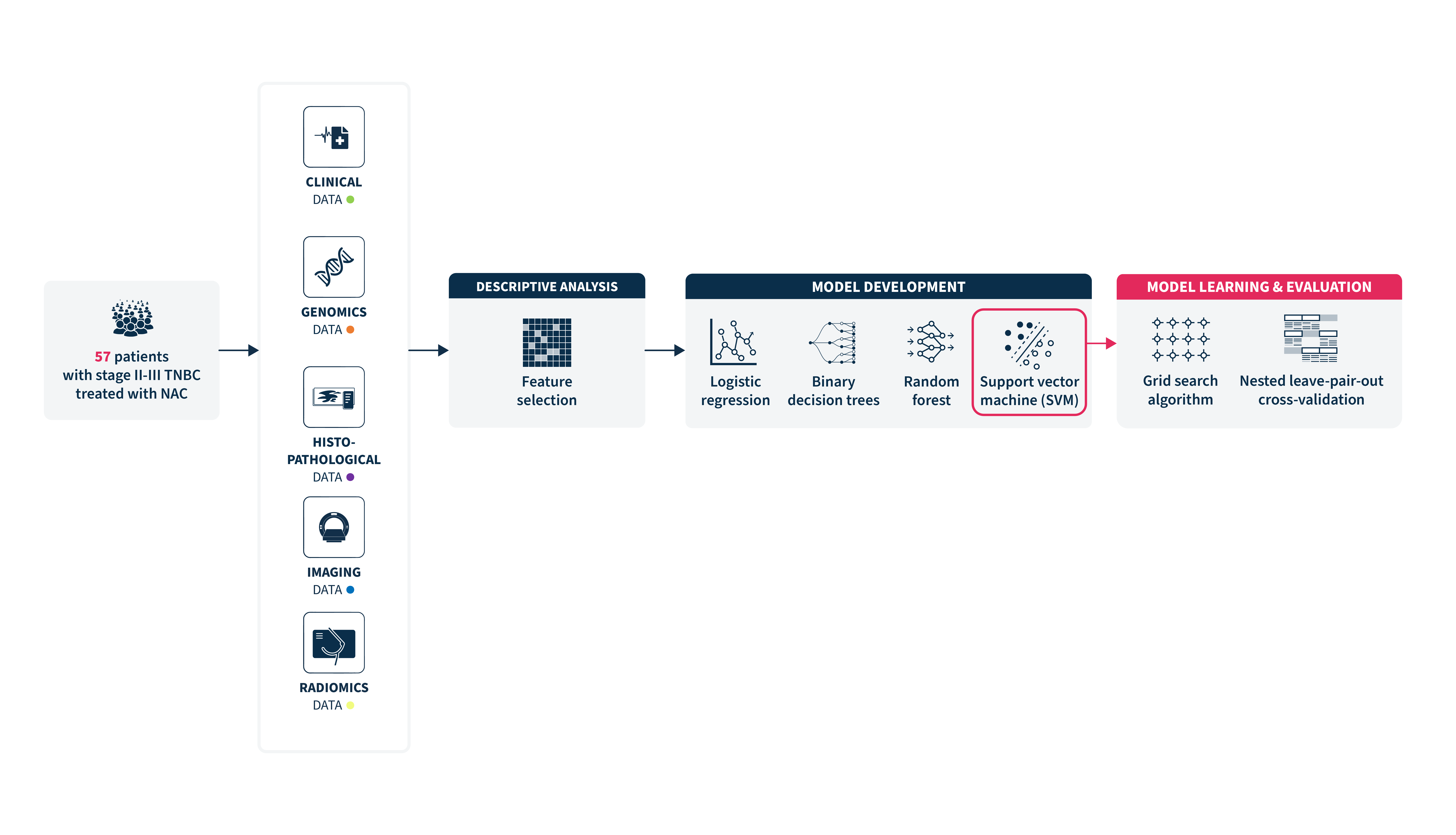
The data collected resulted in 241 predictors per patient, out of which 17 pretherapeutic features were selected for the final analysis.
The best-performing model was a support vector machine (SVM) algorithm with an AUC of 0.82 (95% CI = 0.74-0.90), using the aggregation of all multimodal features (clinical data, histopathological, molecular features, and PET data, including radiomic features). This estimated AUC was significantly greater when considering the entire set of multimodal data, resulting in a 10% decrease in the AUC when a specific data modality was excluded.
The features with the highest weight in the algorithm included a PET-derived SUVmax, two radiomics features, the genomic grade index (GGIr), and clinical T-stage.
Patients predicted to achieve pCR showed an improved event-free survival (EFS) compared to patients with predicted non-pCR. However, due to the small sample size of the study, it requires further research to validate this result.
This study suggests that applying an ML-based method to baseline multimodal data can help predict pCR status after NAC for TNBC patients and may identify correlations with improved long-term outcomes. The integration of clinical, histopathological, molecular, imaging, and radiomic features resulted in a robust AI-powered predictive model.
Identifying patients at greater risk of not achieving pCR earlier could enable a rapid treatment modulation by introducing concomitant treatment with immunotherapy or dose intensification, increasing their chances for better outcomes.
Although the results obtained need an external validation in a large, multicenter population, this study highlighted the importance of adopting a truly multimodal approach to provide better, more efficient, and personalized care to TNBC patients.
Explore this infographic to learn more about this project and the predictive model developed by Groheux et al’s.
Groheux D. et al. Cancers (Basel). 2025. 17(7):1249. doi: 10.3390/cancers17071249.
SOPHiA DDM™ for Radiomics and SOPHiA DDM™ for Multimodal are concepts in development. May not be available for sale.
SOPHiA GENETICS products are for Research Use Only and not for use in diagnostic procedures unless specified otherwise.
SOPHiA DDM™ Dx Hereditary Cancer Solution, SOPHiA DDM™ Dx RNAtarget Oncology Solution and SOPHiA DDM™ Dx Homologous Recombination Deficiency Solution are available as CE-IVD products for In Vitro Diagnostic Use in the European Economic Area (EEA), the United Kingdom and Switzerland. SOPHiA DDM™ Dx Myeloid Solution and SOPHiA DDM™ Dx Solid Tumor Solution are available as CE-IVD products for In Vitro Diagnostic Use in the EEA, the United Kingdom, Switzerland, and Israel. Information about products that may or may not be available in different countries and if applicable, may or may not have received approval or market clearance by a governmental regulatory body for different indications for use. Please contact us to obtain the appropriate product information for your country of residence.
All third-party trademarks listed by SOPHiA GENETICS remain the property of their respective owners. Unless specifically identified as such, SOPHiA GENETICS’ use of third-party trademarks does not indicate any relationship, sponsorship, or endorsement between SOPHiA GENETICS and the owners of these trademarks. Any references by SOPHiA GENETICS to third-party trademarks is to identify the corresponding third-party goods and/or services and shall be considered nominative fair use under the trademark law.