Menu
Menu
Adapted from Dr. Weei-Yuarn Huang’s (Sunnybrook Health Centre, Ontario, Canada) presentation during the webinar “Evaluating Next-Generation Sequencing Solutions for Real-World Clinical Needs in Myeloid Malignancy,” with permission from the author - This concordance study demonstrates the strong analytical performance of the new SOPHiA DDM™ Community Myeloid Solution and its potential to streamline and consolidate myeloid genomic analysis within a single workflow.
About Dr. Weei-Yuarn Huang
Weei-Yuarn Huang is currently the medical director of molecular diagnostics at Sunnybrook Health Science Center in Toronto, Ontario. Under his leadership, the laboratory has become one of largest tumor NGS testing centers in Ontario. He has published more than 50 peer-reviewed manuscripts and served as a member in advisory boards for molecular testing both provincially and nationally.
Acute leukemia classification has progressed from early reliance on morphology and cytochemistry to today’s integrated approach. The WHO classification (now in its 5th edition)1 and the International Consensus Classification2 both emphasize combining morphology, immunohistochemistry, immunophenotyping, cytogenetics, genomic biomarkers and clinical context to achieve accurate diagnosis, inform prognosis, and guide treatment access.
“What has remained the same through all classification updates is the international collaboration and multidisciplinary approach, we need hematopathologists, clinicians, and geneticists integrating clinical context, morphology, phenotyping, and genomic information.”
— Dr. Hubert Tsui
Head, Division of Hematological Pathology, Precision Diagnostics and Therapeutics Program, Sunnybrook Health Science Centre
Across healthcare systems, ensuring equitable and timely access to advanced testing remains a challenge. Variability in laboratory capabilities, bioinformatics pipelines, and reimbursement policies can affect the consistency and timelines of genomic results. Coordinated testing strategies, national guidelines, and advocacy remain essential to ensure every patient benefits from precision diagnostics.
For most patients, the journey toward an acute leukemia diagnosis often begins in the outpatient clinic followed by refereral to an emergency department. The first sign is typically an abnormality in the blood, which may be quantitative (cell counts) or qualitative (cell morphology).
Because acute leukemia care is highly specialized, patients are usually referred to tertiary centers (specialized referral hospitals or cancer centers) for diagnosis and treatment. Many health systems have developed referral frameworks to make this process more consistent.
For example, in Ontario (Canada), the Acute Leukemia Referral Package helps clinicians assess3,4:
One ongoing challenge is deciding where the diagnostic “gold standard” bone marrow aspirate and biopsy should take place: in a community setting, or at a leukemia center. For many community hospitals and even some tertiary leukemia centres, a comprehensive suite of acute leukemia tests may not be available. Given the possibility of additional referred out testing, these logistical elements can impact not only the speed of diagnosis but also the quality and consistency of downstream initial (and reflex) testing, including the most appropriate cytogenetic and molecular investigations.
Bone marrow aspiration (BMA) combined with bone marrow biopsy (BMB) remain the gold standard for diagnosing myeloid neoplasms. Although invasive, it is a bedside procedure that does not require specialized facilities. Multiple tubes are typically drawn at first diagnosis: the initial tube is for morphology, followed by flow cytometry, cytogenetics, and finally one or more tubes for molecular testing.
Historically, this sequence of testing reflected both priority and speed. The diagnosis of acute leukemias relied heavily on morphology and enzyme cytochemistry, as outlined in the French–American–British (FAB) classification system. While foundational, these methods often lacked the precision needed to distinguish biologically and clinically relevant subclasses1,2.
In addition, the conventional tube order can complicate genomic testing, since samples drawn later may be more hemodilute, reducing the analytical sensitivity of downstream molecular analyses (as well as interpretation of variant allele fractions/frequencies).
“By the time you get to tube four or five, is it still bone marrow, or is it blood? What is the actual tumor content that we’re now trying to work up from a genomic perspective?”
— Dr. Hubert Tsui
When acute leukemia is suspected, the need for rapid testing adds further complexity. Not all centers have a full acute leukemia test menu, so samples may need to be distributed across different laboratories, often with extensive paperwork and incomplete electronic health record integration. In some cases, requisitions even arrive with every possible test checked off, creating uncertainty about what to prioritize.
“For some patients, we can’t get a quality aspirate. And there are no or very few circulating leukemia cells. Wow what do we do? ”
— Dr. Hubert Tsui
These practical realities highlight the challenges of ensuring consistent and timely genomic characterization across the spectrum of hematological neoplasms but this is especially important in the context of acute leukemias.
Accurate classification of AML is not only essential for diagnosis but also for access to targeted therapies. Simply labeling a case as acute myeloid leukemia (AML) is no longer sufficient.
Certain biomarkers, such as FLT3-ITD, do not define an AML subtype but act as actionable markers that directly influence treatment. In patients carrying this high-risk mutation, FLT3 inhibitors, such as midostaurin, quizartinib or gilteritinib, are used to improve outcomes5.
Other markers define favorable-risk subtypes, such as t(8;21) or inversion 16. In these cases, the addition of gemtuzumab ozogamicin, an anti-CD33 antibody-drug conjugate6 , to chemotherapy has been shown to improve survival, particularly in patients with core binding factor AML.
These examples underscore how genomic characterization informs both prognosis and therapy selection.
To be clinically actionable, however, results must be delivered quickly. Guidelines recommend turnaround times of 3–5 days for key biomarkers and 5–7 days for cytogenetics, ensuring treatment decisions can be made without delay.
"Just labeling something as acute myeloid leukemia really doesn't suffice anymore"
— Dr. Hubert Tsui
As the AML patient journey becomes increasingly complex, the role of genomic analysis is expanding. Beyond initial diagnosis, classification, and risk stratification, NGS can also inform follow-up, treatment management, and measurable residual disease (MRD) monitoring. Integrating these tools more seamlessly into the clinical workflow will be key to improving outcomes.
This article adapts the presentation of Dr. Hubert Tsui (Sunnybrook Health Centre, Ontario, Canada) during the webinar “Evaluating Next-Generation Sequencing Solutions for Real-World Clinical Needs in Myeloid Malignancy,” with permission from the author.
Hubert Tsui, BSc, MD, PhD, FRCPC
Head, Division of Hematological Pathology, Precision Diagnostics and Therapeutics Program, Sunnybrook Health Science Centre, Associate Scientist, Sunnybrook Research Institute
Dr. Tsui is the division head of hematological pathology at Sunnybrook Health Sciences Centre and an assistant professor in the Departments of Laboratory Medicine and Pathobiology and Immunology at the University of Toronto. He completed his bachelor’s degree, medical degree, doctorate, and residency at the University of Toronto, with a clinical subspecialty in hematological pathology and a research focus on immunology. Tsui leads the Complex Malignant Hematology (CMH) program at Sunnybrook and is the medical-scientific lead of the Sunnybrook Hematology Biobank. He holds leadership roles with the Royal College of Physicians and Surgeons of Canada, Ontario Health – Cancer Care Ontario, and the Canadian Leukemia Study Group. His work focuses on integrating personalized biomarkers into clinical hematopathology and advancing translational research in acute leukemia, MDS, and MPN.
Interested in learning more? Watch the full webinar on demand:
References
Disclaimer
The views expressed in this article are those of the expert and do not necessarily reflect the positions of SOPHiA GENETICS.
Certain products or applications discussed may not be cleared or approved by regulatory authorities in all regions.
All SOPHiA GENETICS products are For Research Use Only. Not for use in diagnostic procedures.
Maslah et al. present a translational study exploring how ruxolitinib and other JAK inhibitors influence clonal evolution in myeloproliferative neoplasms (MPN). Using a SOPHiA DDM™ for Myeloid Malignancies application for evaluation and longitudinal monitoring, the study highlights how treatment pressure can select for RAS pathway mutations, ultimately impacting prognosis and disease progression.

Our infographic distills the study’s key findings, from reduced overall survival to higher transformation risk and the mechanisms driving these effects.
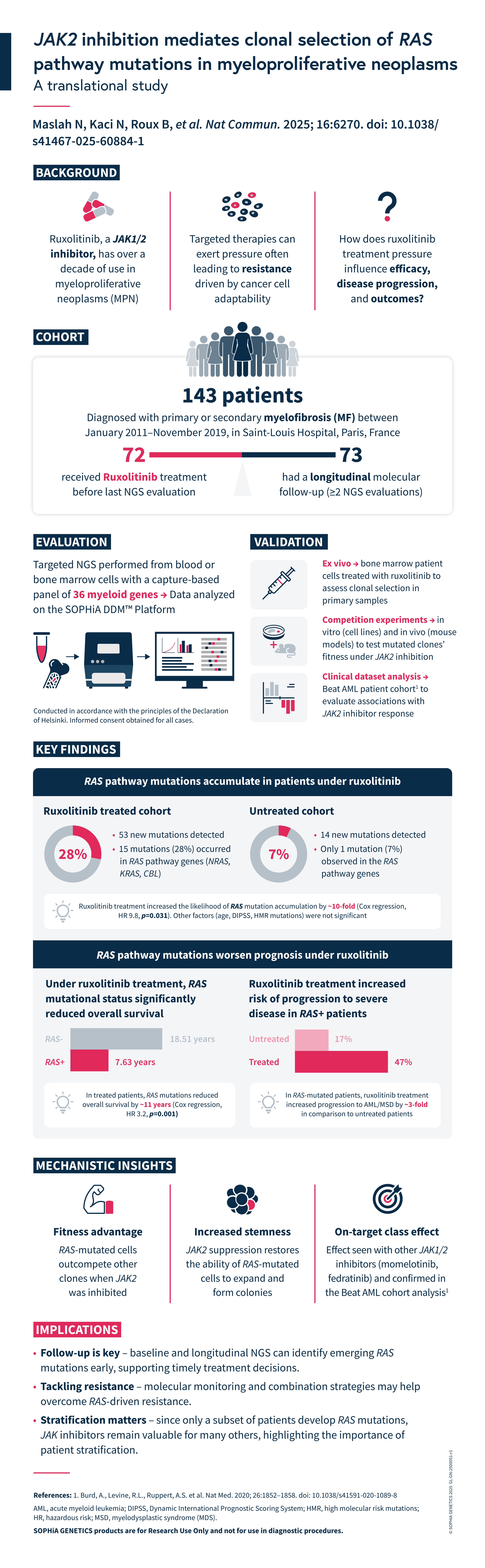
See the full paper for the complete picture.
Maslah N, Kaci N, Roux B, et al. JAK2 inhibition mediates clonal selection of RAS pathway mutations in myeloproliferative neoplasms. Nat Commun. 2025;16:6270. DOI: 10.1038/s41467-025-60884-1
SOPHiA DDM™ for Myeloid Malignancies applications are for Research Use Only.
Table of contents
Gene fusions: what are they and their relevance in myeloid malignancies
Gene fusions occur when two independent genes become abnormally joined, creating a hybrid gene. These events can result from missplicing at the RNA level or structural rearrangements at the DNA level due to damage and faulty repair processes. Such rearrangements often include chromosomal translocations, interstitial deletions, and inversions1
The resulting fusion involves a driver gene and one or more partner genes, joined at specific breakpoints that give rise to different fusion isoforms. These hybrid genes often encode abnormal fusion proteins that can hijack key cellular processes and promote oncogenesis.
A landmark example is the Philadelphia chromosome in chronic myeloid leukemia (CML), a translocation between chromosomes 9 and 22 that produces the BCR::ABL1 fusion gene. This discovery transformed CML treatment and paved the way for targeted therapies such as imatinib, a tyrosine kinase inhibitor (TKI) that dramatically improved patient survival[1].
Since then, numerous other fusion genes involving tyrosine kinases have been identified in myeloproliferative malignancies, including:
These findings define a subgroup of disorders now classified as myeloid neoplasms with eosinophilia and abnormalities in PDGFRA, PDGFRB, or FGFR1. But while fusions involving PDGFRA and PDGFRB respond well to imatinib, FGFR1-driven diseases do not, highlighting the critical need for accurate and timely fusion detection2.
Beyond tyrosine kinase fusions, several other fusion genes play critical roles in blood cancers and carry important diagnostic and prognostic implications3:
Traditional cytogenetic techniques, such as chromosomal banding, alongside molecular methods like fluorescence in situ hybridization (FISH), gene fusion microarrays, and PCR/RT-PCR, were among the earliest clinical assays developed to detect fusion genes.
Today, these approaches remain highly sensitive and are routinely used for orthogonal confirmation of fusion findings, helping ensure diagnostic accuracy. However, they remain hard to scale due to their inherent limitations:
While still indispensable in many diagnostic workflows, these methods are increasingly being complemented by next-generation sequencing (NGS) technologies.
NGS-based approaches have dramatically expanded the scope of fusion detection, uncovering previously undetectable events and enabling a more comprehensive view of the genomic landscape. When integrated with traditional methods, these advanced tools help streamline diagnostics and improve clinical decision-making in the context of myeloid malignancies5.
Targeted capture panels based on DNA or RNA offer a focused and efficient approach to detecting gene fusions. These panels rely on custom-designed probes that target regions commonly involved in fusion breakpoints1,2,5.
In DNA-based NGS, probes aim to capture intronic regions where fusion breakpoints often occur. In this case, probe design and bioinformatic analysis pipelines play a key role in the sensitivity of the detection. Since fusion breakpoints frequently lie in large intronic regions that are GC-rich, repetitive, and structurally complex, this can impact capture efficiency and analysis complexity.
Despite these challenges, DNA-based capture panels remain valuable for:
RNA-based NGS, by contrast, offers several advantages. Rather than capturing introns, RNA methods detect exon–exon junctions, which directly indicate the presence of expressed fusion transcripts. This simplifies probe or primer design and improves detection in regions that are difficult to sequence from DNA. RNA-based approaches are generally more sensitive than DNA-based ones and are especially useful for identifying expressed, clinically relevant fusions, including those with unknown or variable breakpoints.
However, RNA-based fusion detection is not without challenges:
Despite these challenges, RNA sequencing remains a powerful tool, often used in tandem with DNA-based methods to increase confidence in fusion calls. In practice, DNA- and RNA-based approaches are complementary. DNA sequencing enables broader biomarker profiling from stable material, while RNA provides functional confirmation and increased detection sensitivity. Together, they support a more comprehensive understanding of gene fusions in myeloid malignancies.
DNA-based fusion detection within the SOPHiA DDM™ Platform is driven by a two-pronged strategy that combines optimized probe design with advanced algorithmic analysis:
CARDAMOM pinpoints potential fusion events (also referred to as adjacencies) by analyzing two key signals in sequencing data:
Once key read signals are detected, CARDAMOM clusters them into candidate breakpoints and applies a rigorous multi-step process to ensure only high-confidence fusions are reported. This fusion-calling workflow is designed to maximize sensitivity while maintaining analytical precision.
Discover how our comprehensive application, SOPHiA DDM™ Community Myeloid Solution, enables confident fusion detection in clinical samples.
Webinar: Evaluating Next-Generation Sequencing Solutions for Real-World Clinical Needs in Myeloid Malignancy → Watch the webinar
Visit our SOPHiA DDM™ Community Myeloid Solution page to learn more about this application.
References
More than six people die every hour in the US from a blood cancer. Solutions can’t come fast enough for those who suffer with these cancers all around the world. Fortunately, researchers studying blood diseases have experienced rapid advances in their capabilities to develop and test effective therapies with some extremely significant advancements.
Some of the most difficult limitations of molecular profiling for hematological cancer disorders include accurate detection of mutations in GC-rich gene regions and insertions or deletions in challenging genes. Data analysis on NGS DNA samples identifies complex variants to accurately identify myeloid malignancies. This validation of targeted mutations has encouraged many medical centers to order NGS testing for every acute myeloid leukemia case.
Faster, more efficient NGS analysis can drive better hematological cancer research outcomes to potentially improve care for patients with blood cancers and diagnosis of new cases.
International guidelines for hematological cancer diagnosis and treatment are continuously evolving and create the need for laboratories’ fast adaptation. Those evidence-based guidelines by physician commissions contribute to improving the clinical standard of care. The World Health Organization, European Hematology Association, European LeukemiaNet, College of American Pathologists and the American Society of Hematology call for increased use of NGS testing for initial diagnostic workup of blood cancers.
Detection of the relevant biomarkers for myeloid malignancies by NGS, per international guidelines, helps to ensure optimal clinical trial enrollment, therapy validation, dose protocols and other research benefits. A solution that can be constantly updated and inform based on those guidelines ensures that the research is always current.
The accurate assessment of biomarkers and the validity of resulting research findings depend on reliable DNA and RNA fusion panels and easily reproducible results. Data analysis and reporting in a comprehensive platform eliminates silos of valuable data and maximizes its application.
The SOPHiA DDM™ Platform enables the upload of multimodal data from any environment to one of the world’s largest networks of connected labs. Data remains the property of the healthcare institution, but pseudonymized and pooled with like data, it can propel research and ultimately treatment forward with the goal of improved patient care.
Learn more about the capabilities of SOPHiA DDM™ Platform for myeloid biomarker detection and more by contacting us today.
SOPHiA DDM™ Community CLL Clonality Solution is an all-in-one application developed and tested by European Hemato-Oncology experts within the SOPHiA GENETICS Community for the analysis and stratification of CLL cases. This sample-to-report approach combines an optimized next-generation sequencing (NGS) workflow and the powerful analytics of the SOPHiA DDM™ Platform.
In this study, we demonstrate the effectiveness of the SOPHiA DDM™ application in generating a comprehensive report on the genomic status of CLL in a fictitious case. Download the application note and learn more about the SOPHiA DDM™ Community Clonality Solution CLL.
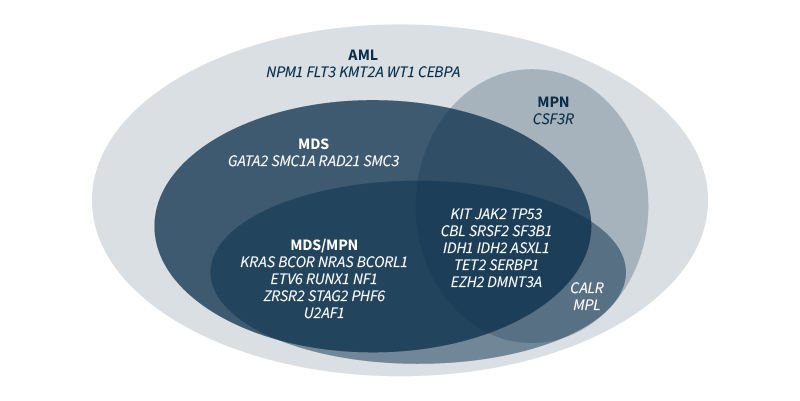
Myeloid malignancies or neoplasms are rare clonal disorders of hematopoietic stem or progenitor cells. They are caused by genetic and epigenetic changes that disrupt fundamental cellular processes, such as self-renewal, differentiation, and proliferation.1 Mutations encountered in myeloid malignancies include somatic variants, such as single nucleotide variants (SNVs), insertions and deletions (Indels), gene fusions, and copy number variations (CNVs). Some of the genetic changes are strongly associated with a specific phenotype (e.g., BCR/ABL fusion in chronic myeloid leukemia), but most are not linked to a single diagnostic entity (Figure 1).2
Accurate detection of mutations is extremely important for the diagnosis and prognosis of myeloid malignancies, and to identify the genomic events that are classified as ‘actionable’, i.e., those for which targeted therapies exist. The advent of next generation sequencing (NGS)-based techniques greatly enhanced our capability to detect mutations. There are numerous advantages of using NGS: it is a high-throughput method that allows for massive parallel sequencing of multiple targeted genomic regions in many samples, shortening the time to report. Moreover, differently to traditional methods, an NGS analysis requires small amounts of tumor DNA/RNA. However, NGS has also brought about some challenges.
Mutations and their patterns in myeloid malignancies are heterogeneous and must be identified, warranting the use of NGS for their detection. Ensuring that a gene panel is comprehensive enough to detect all variants implicated in myeloid neoplasms is one of the biggest challenges for a clinical laboratory performing molecular testing of new or suspected cases.2 Compiling a gene list is a tall order that requires a careful analysis of recent publications, FDA-approved targeted therapies, scientific society guidelines, and the products and services offered by NGS companies. A balance is needed – a very large gene panel will be expensive to run and time-consuming to analyze, whilst a small panel may miss important information.
SOPHiA GENETICS offers offers expertly designed applications with key genes associated with hematological malignancies, and also supports healthcare institutions in the design of custom applications with the desired gene content to fully meet their research needs.
Three genes are notoriously difficult to assess by NGS in myeloid neoplasm samples: FLT3, CEBPA, and CALR. Challenges with FLT3 internal tandem duplication (ITD) mutation detection derive from difficulties in aligning sequence reads with large duplications. Moreover, larger duplications are missed by amplicon-based panels, if the sequencing read is shorter than the ITD size. As FLT3-ITD length has a prognostic value, the identification of the ITD must be precise. False negatives in CEBPA mutation detection are often encountered in assays that are not carefully designed and validated, whereas CALR is problematic in NGS due to difficulties in detecting Indels resulting in frameshift mutations, e.g., a common 52bp deletion variant.2 Complex genes require optimal probe design, coverage and analytic pipelines.
Complex genes require optimal probe design, coverage and analytic pipelines.
The SOPHiA DDM™ for Blood Cancers portfolio has been designed to offer uniform coverage of complex genomic variants to ultimately allow:
A timely identification of mutations is of crucial importance for two genes in particular: TP53 and FLT3. A mutation in TP53 correlates with chemoresistance and identifies patients in whom standard therapy will do more harm than good. Timely identification of FLT3-ITD is needed to introduce a recently approved treatment with a specific kinase inhibitor that must be administered on day 8 of the induction therapy.2 However, the average laboratory turnaround time for NGS assays is 13 to 21 days, which makes NGS-based FLT3 detection unfeasible for first-line treatment decisions.2 To solve this challenge, continuous optimization of all aspects of the laboratory workflow is needed.
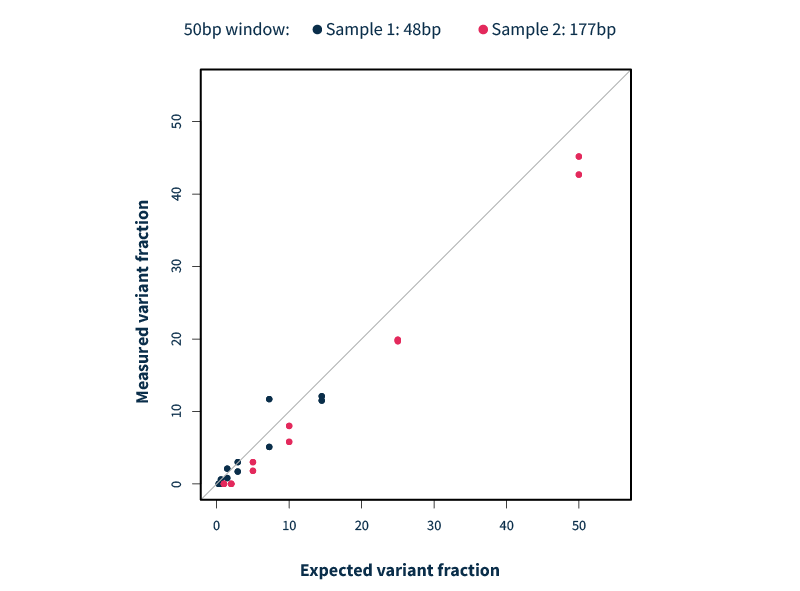
The advanced analytics of SOPHiA DDM™ provide optimized variant detection in a single workflow. For example, the SOPHiA DDM™ for Blood Cancer applications have been trained to efficiently detect all FLT3 fragment sizes, irrespective of the insertion point. All confirmed ITDs were detected (no false negatives) and no additional variant was called (no false positives), making it a robust and reliable detection method.
In addition to the technical challenges described above, a laboratory performing NGS-based mutation profiling in myeloid neoplasms faces several variant interpretation problems. NGS techniques result in a huge number of genetic variants needing interpretation, which requires a significant amount of time and expertise from institutions. Also, many genes may acquire novel mutations that are not present in NGS databases. Often, such mutations are classified as variants of uncertain significance and are of little value for the prognosis and guidance on therapeutic choices. Computational prediction algorithms can help institutions identify and understand biologically relevant, actionable biomarkers. All such analyses should be incorporated into the bioinformatic pipeline to produce the end report for the clinician.3
Computational prediction algorithms can help institutions identify and understand biologically relevant, actionable biomarkers
With the SOPHiA DDM™ Platform, experts can flag the pathogenicity level of variants. Flagging decisions are supported by access to SOPHiA GENETICS’ global community of knowledge sharing and qualified databases containing relevant information on the variants, including those of unknown significance.
SOPHiA GENETICS products are for Research Use Only and not for use in diagnostic procedures unless specified otherwise.
SOPHiA DDM™ Dx Hereditary Cancer Solution, SOPHiA DDM™ Dx RNAtarget Oncology Solution and SOPHiA DDM™ Dx Homologous Recombination Deficiency Solution are available as CE-IVD products for In Vitro Diagnostic Use in the European Economic Area (EEA), the United Kingdom and Switzerland. SOPHiA DDM™ Dx Myeloid Solution and SOPHiA DDM™ Dx Solid Tumor Solution are available as CE-IVD products for In Vitro Diagnostic Use in the EEA, the United Kingdom, Switzerland, and Israel. Information about products that may or may not be available in different countries and if applicable, may or may not have received approval or market clearance by a governmental regulatory body for different indications for use. Please contact us to obtain the appropriate product information for your country of residence.
All third-party trademarks listed by SOPHiA GENETICS remain the property of their respective owners. Unless specifically identified as such, SOPHiA GENETICS’ use of third-party trademarks does not indicate any relationship, sponsorship, or endorsement between SOPHiA GENETICS and the owners of these trademarks. Any references by SOPHiA GENETICS to third-party trademarks is to identify the corresponding third-party goods and/or services and shall be considered nominative fair use under the trademark law.