Menu
Menu
Unlock the future of precision medicine with cutting-edge SOPHiA DDM™ technology powering NGS workflows, delivering unparalleled accuracy and ease.
Our pharmacogenomics applications were developed in collaboration with leading PGx experts to encompass CPIC and FDA recommendations. They quickly and accurately identify PGx-associated genomic variants of clinical significance, powered by the advanced analytics of the SOPHiA DDM™ Platform.

CNVs are accurately detected by our MUSKAT™ algorithm alongside SNVs and Indels in a single experiment capable of detecting known and previously undetected variants.


Major haplotypes (star alleles) for VIP genes according to PharmVar and PharmGKB are assigned and annotated with phenotypic consequences by our AI agent, STAR ANISE.


CNVs, star alleles, and promoters/UTRs in CYP2D6 are accurately called, including information for CYP2D6/CYP2D7 pseudogene analysis.

Boost lab efficiency with an end-to-end pharmacogenomics NGS workflow. Perform next-generation sequencing in house, or leverage SOPHiA DDM™ Integrated Access Mode to tap into a global network of sequencing labs. All variant detection and interpretation steps are powered by the SOPHiA DDM™ Platform, giving you full control over your data and samples.

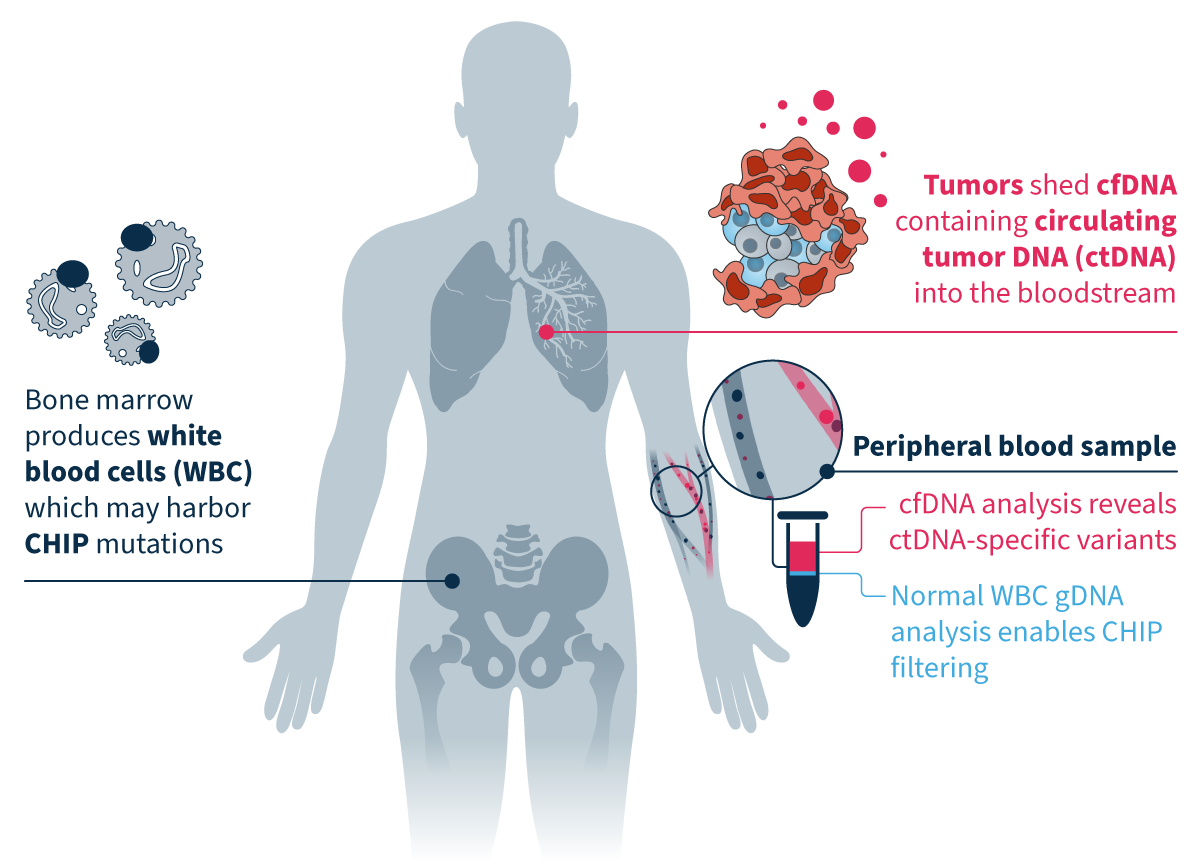
By sequencing tumor-derived cfDNA and normal white blood cell DNA in parallel, you can effectively reduce noise in your data analysis and focus on tumor-specific somatic variants.
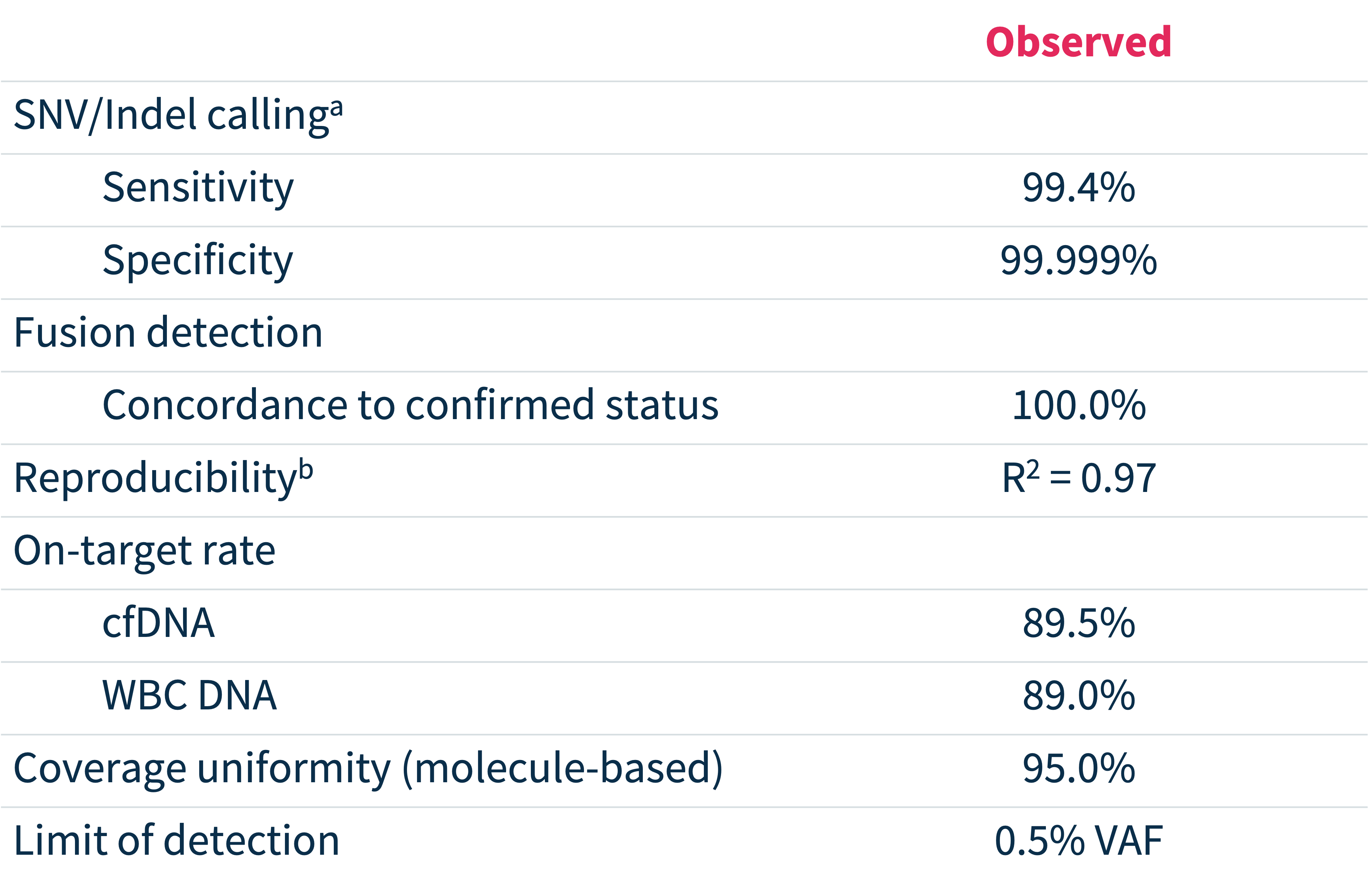
Exceptional analytical performance3
PPA, positive percent agreement; VAF, variant allele frequency. a Based on end-to-end concordance analysis between MSK-ACCESS® powered with SOPHiA DDM™ and centralized version of MSK-ACCESS® at MSK, using 48 clinical cfDNA + matched WBC DNA samples; b Correlation of variant allele fraction to reference method.

Traditional PGx testing techniques, such as PCR and arrays, can be labor-intensive and limit the discovery of novel variants. NGS on the other hand, can assess all relevant variants within your region of interest, consolidating multiple workflows into a single experiment, with long-range PCR as a complementary option to resolve ambiguous results.

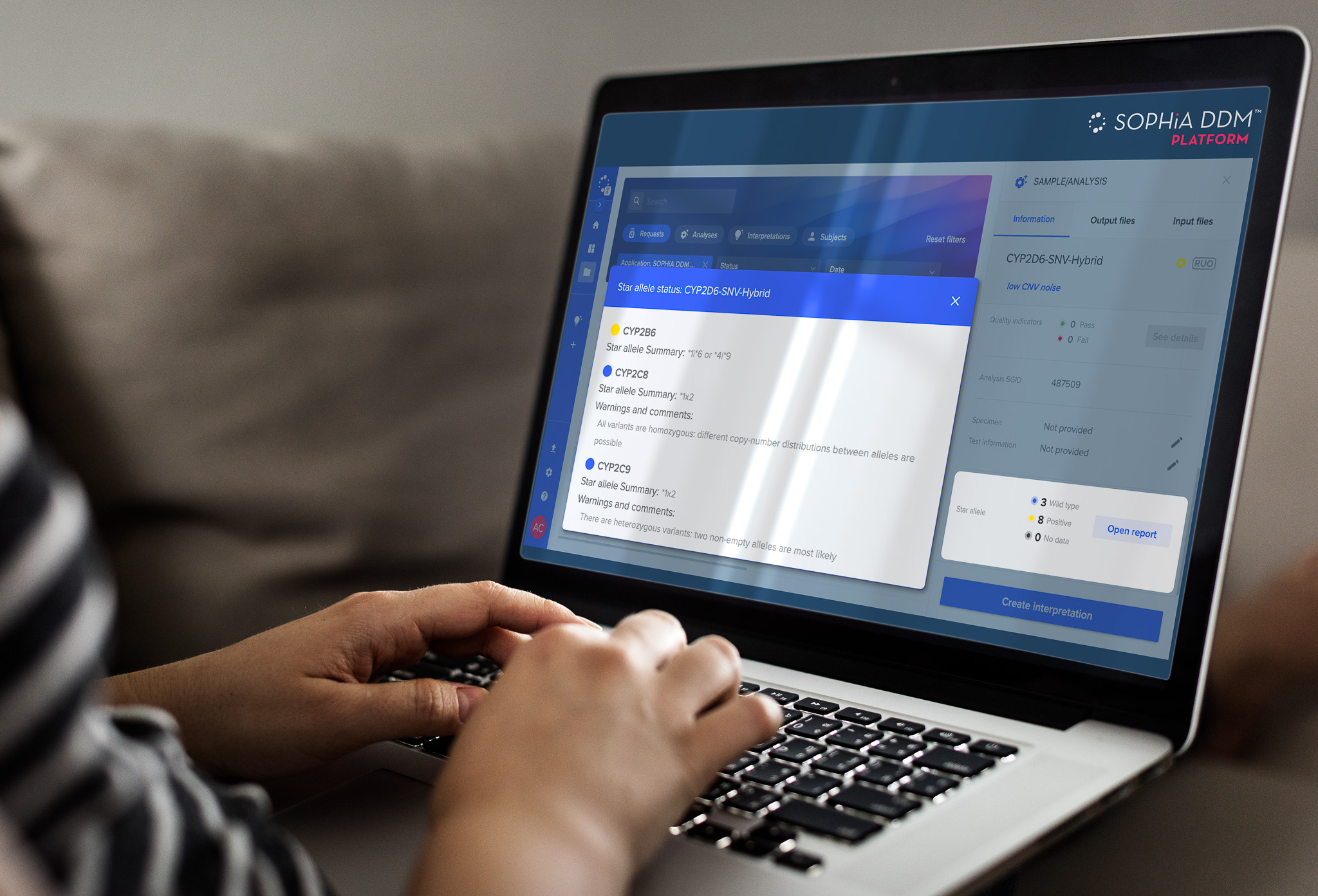
The STAR ANISE AI agent phases variants across 11-15 PharmVar-curated genes, identifies major haplotypes (star alleles), and annotates them with phenotypic consequences based on CPIC guidelines. STAR ANISE achieves 100% concordance with ground-truth CYP2D6 star allele calling, while SOPHiA DDM™ presents results in a clear, intuitive interface, enabling rapid, confident review of complex variant data.
| SOPHiA DDM™ Community Pharmacogenomics Solution | SOPHiA DDM™ Community Extended Pharmacogenomics Solution | |
|---|---|---|
| Research applications | Oncology, cardiovascular diseases, psychiatric disorders, pain management, infectious diseases, epilepsy, gastrointestinal diseases | |
| Content | 42 genes including a pseudogene | 74 genes including a pseudogene, HLA, and mtDNA coverage |
| Variants called | SNVs, Indels, CNVs, star alleles (11 genes), CYP2D6/CYP2D7 gene conversion | SNVs, Indels, CNVs, star alleles (15 genes), CYP2D6/CYP2D7 gene conversion |
| Sample type | Blood | Blood |
| Starting material | 50 ng DNA | 50 ng DNA |
| Reads per sample | 2 million | 4 million |
| Panel size | 77 kb | 260 kb |
| Multiplexing recommendations (samples per run)* |
8 and 24 for Illumina MiniSeq™ Mid- and High-Output Kits 16 and 24 for Illumina MiSeq® v2 and v3 96 and 384 for Illumina NextSeq® 550 Mid- and High-Output Kits |
12, 24, and 48 for Illumina Miseq™ i100 25M, 50M, and 100M flowcell 12 for Illumina MiSeq® v3 48 and 192 for Illumina NextSeq™ 500/550 Mid- and High-Output Kits 48, 192, 576, and 864 for Illumina NextSeq™ 1000/2000 P1, P2, P3, and P4 |
| Library prep and capture hands-on time + FASTQ analysis time | 5 hours + 2 hours | 5 hours + 3 hours |
| *When working with more than 100 Gb or 96 samples, data upload to the SOPHiA DDM™ Platform needs to be carried out with the SOPHiA DDM™ Command Line Interface (CLI) Tool. | ||
SOPHiA GENETICS products are for Research Use Only and not for use in diagnostic procedures unless specified otherwise.
SOPHiA DDM™ Dx Hereditary Cancer Solution, SOPHiA DDM™ Dx RNAtarget Oncology Solution and SOPHiA DDM™ Dx Homologous Recombination Deficiency Solution are available as CE-IVD products for In Vitro Diagnostic Use in the European Economic Area (EEA), the United Kingdom and Switzerland. SOPHiA DDM™ Dx Myeloid Solution and SOPHiA DDM™ Dx Solid Tumor Solution are available as CE-IVD products for In Vitro Diagnostic Use in the EEA, the United Kingdom, Switzerland, and Israel. Information about products that may or may not be available in different countries and if applicable, may or may not have received approval or market clearance by a governmental regulatory body for different indications for use. Please contact us to obtain the appropriate product information for your country of residence.
All third-party trademarks listed by SOPHiA GENETICS remain the property of their respective owners. Unless specifically identified as such, SOPHiA GENETICS’ use of third-party trademarks does not indicate any relationship, sponsorship, or endorsement between SOPHiA GENETICS and the owners of these trademarks. Any references by SOPHiA GENETICS to third-party trademarks is to identify the corresponding third-party goods and/or services and shall be considered nominative fair use under the trademark law.