SOPHiA DDM™ Hereditary Cancer Solutions
Confidently assess genetic variants predisposing to cancer
Accurately characterize the complex mutational landscape associated with major hereditary cancer disorders.
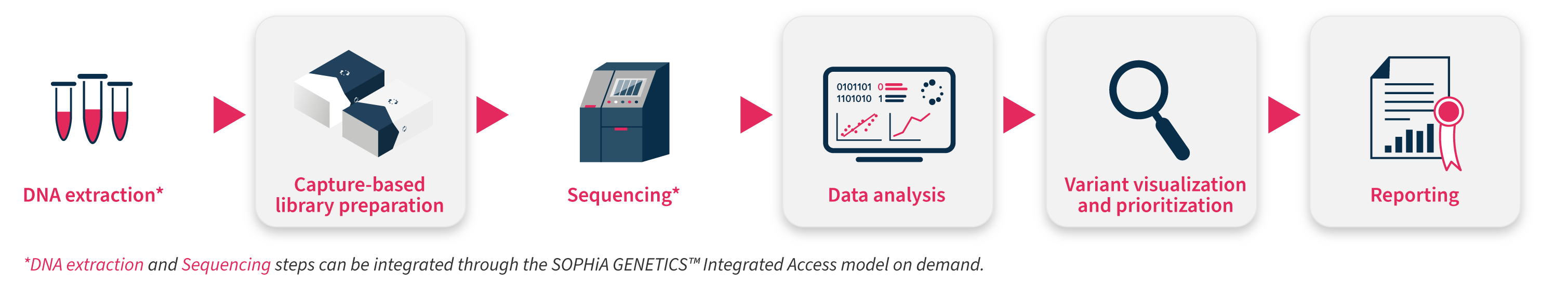
All our next generation sequencing (NGS)-based applications have been conceived to overcome sequencing bias and maximize performance, combining an expertly designed capture-based workflow with the analytical capabilities and interpretation-support functionalities of the SOPHiA DDM™ Platform. Our pre-designed applications help you increase the efficiency of your laboratory, offering an end-to-end approach (from sample to variant report) to turn high-quality data into valuable insights.
Announcing SOPHiA DDM™ Hereditary Cancer Solution (HCS) v2.0!
Enhanced with expanded, guideline-driven content and new features to accelerate analysis.
Ready-to-sequence target enriched library in just 1.5 days
Optimized automation protocols for a variety of liquid handling robots to support high-throughput analyses
Customizable content to meet your specific research laboratory needs
Accurate detection and annotation of challenging variants, including SNVs, long Indels and CNVs in a single assay
Streamlined interpretation through intuitive variant filters, machine learning-based variant classification (complementing the ACMG ranking), and access to one of the largest networks of connected healthcare institutions to gain and share knowledge on relevant variants
Product Details
Automated workflow to increase your productivity
The SOPHiA DDM™ Hereditary Cancer Solution (HCS) can be coupled with leading liquid handling robots for a fully automated library preparation. As a result, you can benefit from a standardized workflow, that provides high quality libraries for reliable sequencing while also increasing sample throughput and decreasing required hands-on time to as little as 1.45 hours.
Performance
|
Observed |
Lower 95% CI |
|---|---|---|
| Sensitivity | 100% | 99.20% |
| Specificity | 100% | 99.99% |
| Accuracy | 100% | 99.99% |
| Precision | 99.86% | 96.42% |
| Repeatability | 99.98% | 99.98% |
| Reproducibility | 99.93% | 99.93% |
Values have been calculated for SNVs and Indels only from a total of 159 samples processed on MiSeq®
Analysis based on SOPHiA DDM™ HCS v1.1.
Deeply assessed to not miss any variant
The SOPHiA DDM™ HCS accuracy has been assessed through a multicenter performance evaluation study on 159 samples with 373 unique variants coming from 7 sequencing centers. The study showed:
- High on-target rates and coverage uniformity
- High-confidence calling of SNVs, Indels and CNVs in all genes of the panel
- Reliable detection of complex variants, such as Alu insertions
- PMS2 and PMS2CL variants
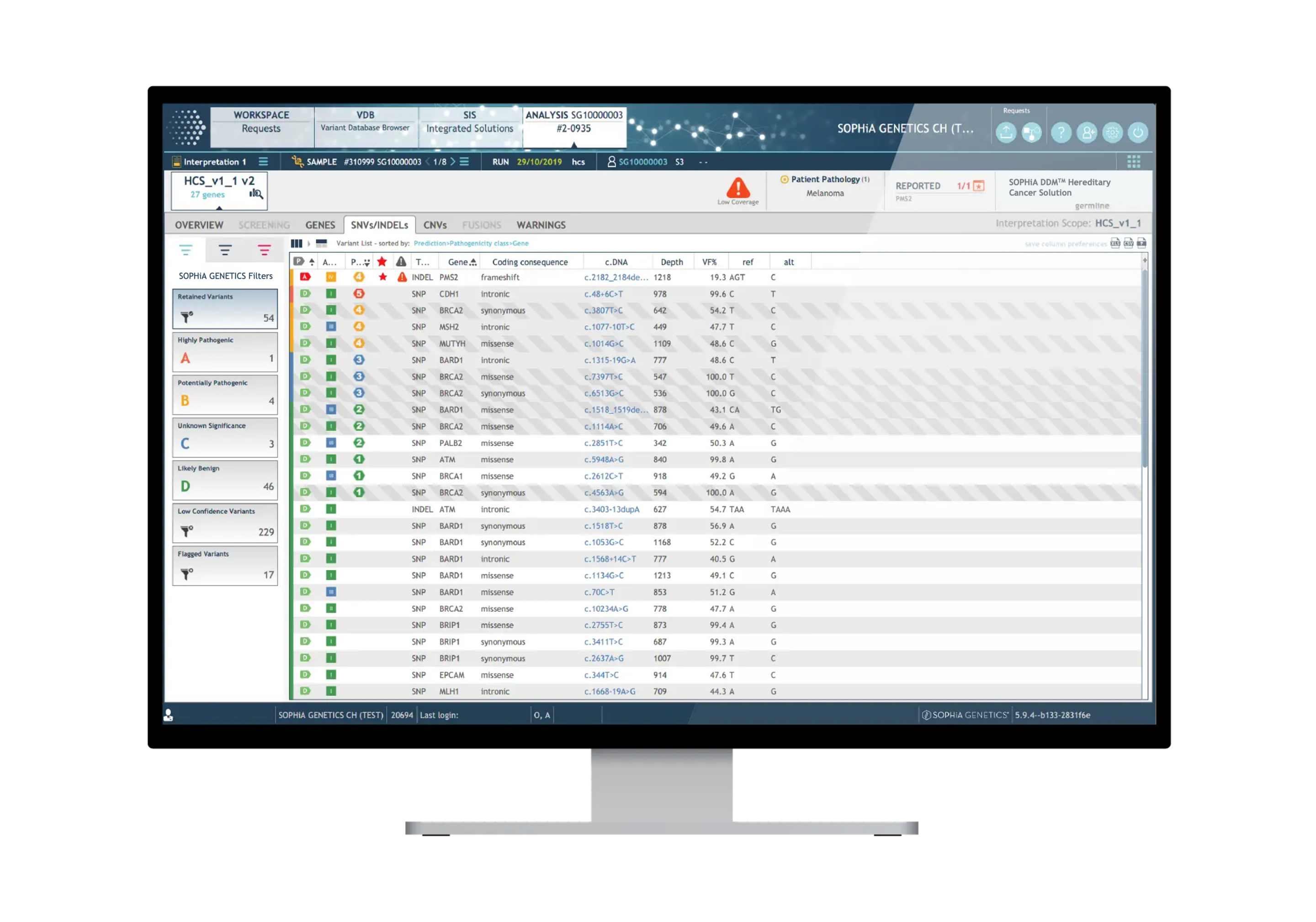
Dedicated features to ease variant interpretation
The SOPHiA DDM™ Platform features intuitive variant filters and prioritization options to streamline the interpretation process and help you significantly reduce turnaround time.
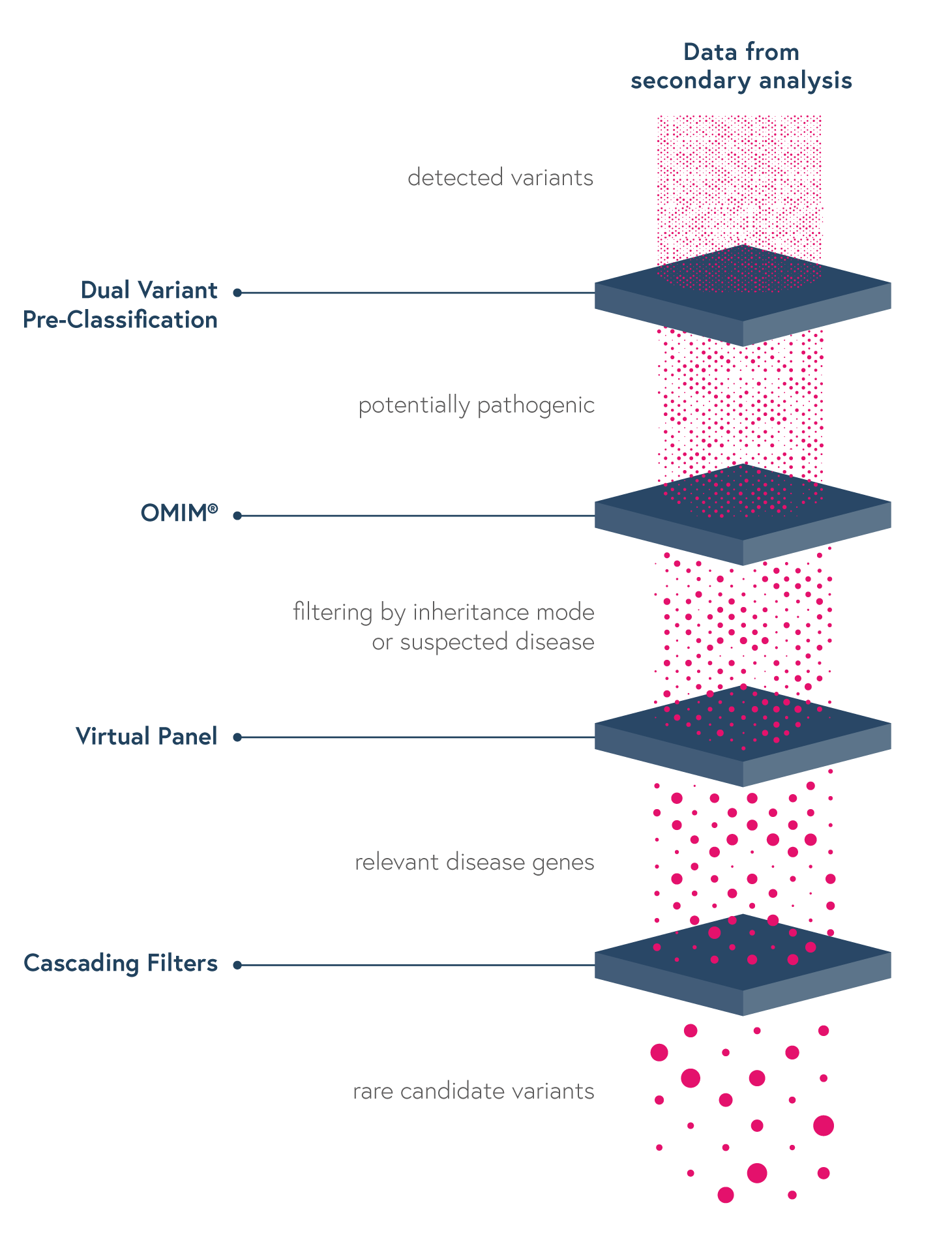
• Dual Variant Pre-classification to improve assessment of variants pathogenicity based on both ACMG scores and our machine learning predictions
• Virtual Panels to restrict the interpretation to sub-panels of genes using the HPO or OMIM browser
• Cascading Filters to apply custom filtering options for quicker screening of relevant variants and save strategies for future analyses
Through SOPHiA DDM™, you can also have access to Alamut™ Visual Plus, a fullgenome browser that integrates numerous curated genomic and literature databases, guidelines, missense and splicing predictors, thus enabling a deeper variant exploration.
Specifications
| Parameters | SOPHiA DDM™ HCS v1.1 |
SOPHiA DDM™ HCS v2.0 |
Hereditary Cancer Community Panels Selected panels – contact us for more information |
|||
|---|---|---|---|---|---|---|
| Diseases Covered | Hereditary Breast and Ovarian Cancer (HBOC), Lynch and various intestinal polyposis syndromes | Breast, ovarian, prostate, abdominal, endocrine & neuroendocrine, nervous system, renal, and skin | Breast, ovarian, prostate, Lynch Syndrome, polyposis, rare CRC syndromes, gastric, pancreatic, cutaneous tumors, melanoma, non-melanoma skin cancer, renal, thyroid, neuroendocrine, nervous system, fanconi anemia, sarcoma | |||
| Genes | 26 + PMS2CL | 82 + PMS2CL | 37 + PMS2CL | 66 | 143 | 117 |
| Target Region Size | 105 kb | 285kb | 92 kb | 222 kb | 405 kb | 371 kb |
| Sample Type | Blood | Blood | Blood | Blood | Blood | Blood |
| DNA Input Amount | 200 ng | 50 ng | 200 ng | 200 ng | 200 ng | 200 ng |
| Sequencer Compatibility |
|
|
|
|
|
|
| Library Preparation Time | 1.5 days | 1.5 days | 1.5 days | 1.5 days | 1.5 days | 1.5 days |
| Analysis Time From FASTQ | 4 hours | 4 hours | 4 hours | 4 hours | 4 hours | 4 hours |
| Detected Variants |
|
|
|
|
|
|
Resources
Related webinars
Read more
Contact us
Please fill out the form below to get in touch
Related products
SOPHiA DDM™ For Solid Tumors
From targeted to comprehensive genomic profiling, our solutions support healthcare professionals in their journey to analyze solid tumors.
SOPHiA DDM™ For Blood Cancers
Our future-proof end-to-end solutions allows detection and characterization of complex genomic variants associated to different blood cancer disorders.