As the list of biomarkers and disease-associated genes expands, so does the number of necessary molecular testing that a laboratory has to undertake.
Given the constant progress in targeted therapy and the expanding lists of international recommendations, it is nearly impossible to keep pace using traditional single-gene testing methods. Tailored NGS-based workflows empower specialists to get high-quality and reproducible data on up-to-date gene panels to accelerate their studies.
Expertly designed gene panels customizable to meet your laboratory’s specific needs
Detection of challenging variants including SNVs, Indels, CNVs, fusions, and tandem duplications in KMT2A and FLT3
Tailored probes to maximize on-target rate and coverage uniformity, for a cost-effective multiplexing
Ready-to-use target-enriched library in just 2 days
Data analysis from FASTQ files in as little as 4 hours
Streamlined workflow, from sample to report, to accelerate your analysis
Easy variant visualization, classification and annotation within the SOPHiA DDM™ Platform
Tertiary analysis based on the latest scientific evidence on relevant variants with OncoPortal™Plus
Access to SOPHiA GENETICS Community, one of the largest networks of connected healthcare institutions, to gain and share scientific knowledge
Accurately characterize myeloid malignancies
Myeloid neoplasms originate from hematopoietic disruptions in the myeloid lineage due to highly heterogeneous and complex mutations. Improvements in sequencing technics and the development of targeted therapies call for a refined classification according to genetic alterations1,2,3. Therefore, advances in cancer study depend on timely, cost-effective, and reliable high-throughput sequencing strategies.
Myelodysplastic syndromes (MDS)
Their classification still highly depends on the blast count. However, some genetic abnormalities determine subclasses of MDS such as 5q deletions, SF3B1 mutations, or TP53 biallelic inactivation1.
Myeloproliferative neoplasms (MPN)
They include, but are not restricted to, Chronic myeloid leukaemia (CML), Polycythaemia vera (PV) and Essential thrombocythaemia (ET) for which most patients exhibit mutations in JAK2, CALR, or MPL genes2. Other mutations in ASXL1 and EZH2 have been observed1.
Acute myeloid leukemia (AML)
Chronic myeloid leukemia (CML)

Confidently assess challenging biomarkers
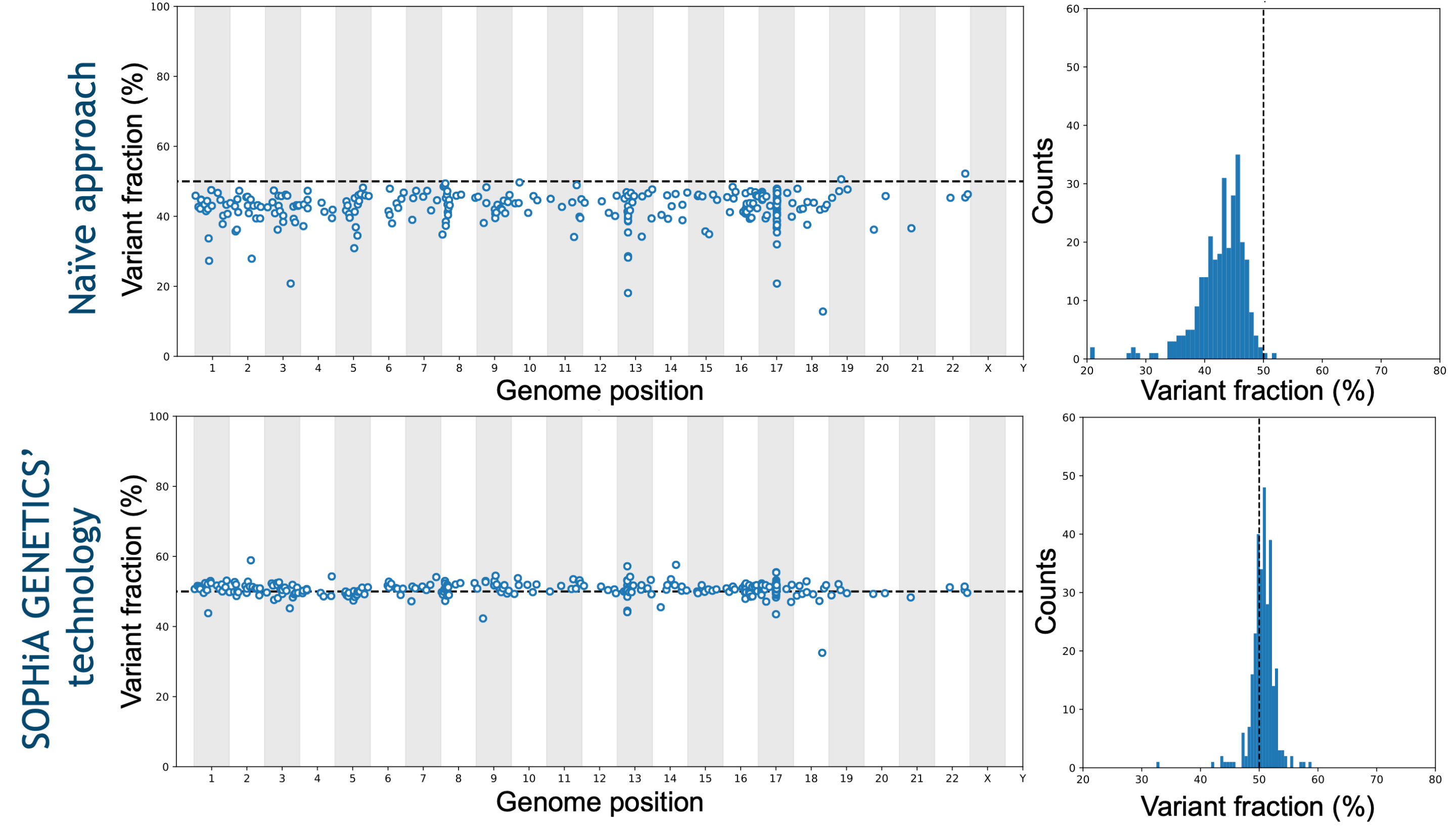
Molecular profiling by NGS has introduced a paradigm shift in investigating the pathogenic variants causing different myeloid malignancies. Indeed, it allows the characterization of multiple SNVs, Indels, CNVs and fusions in one unique experiment. With an optimized probe design and the advanced analytical capabilities of the SOPHiA DDM™️ Platform, challenging variants in GC-rich regions such as ASXL1, CALR, CEBPA, and tandem duplications in KMT2D and FLT3 are covered.
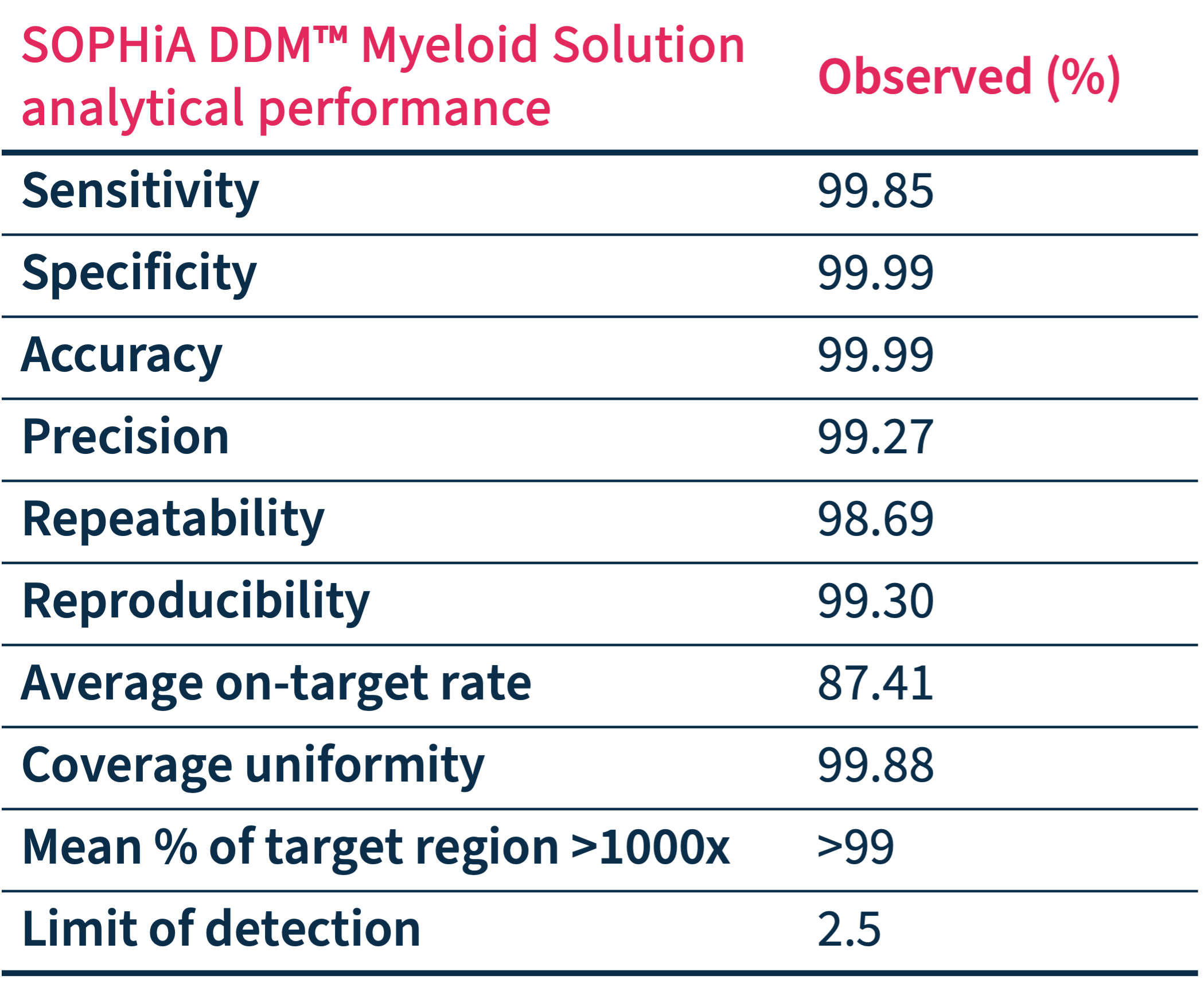
Trust your data
Exceptional Analytical Performance
Addressing the technical limitations related to the analyses of key biomarkers, the SOPHiA DDM™️ Myeloid Solution and its derived applications combined with the SOPHiA DDM™️ Platform exhibit advanced analytical capabilities.
Data on file.
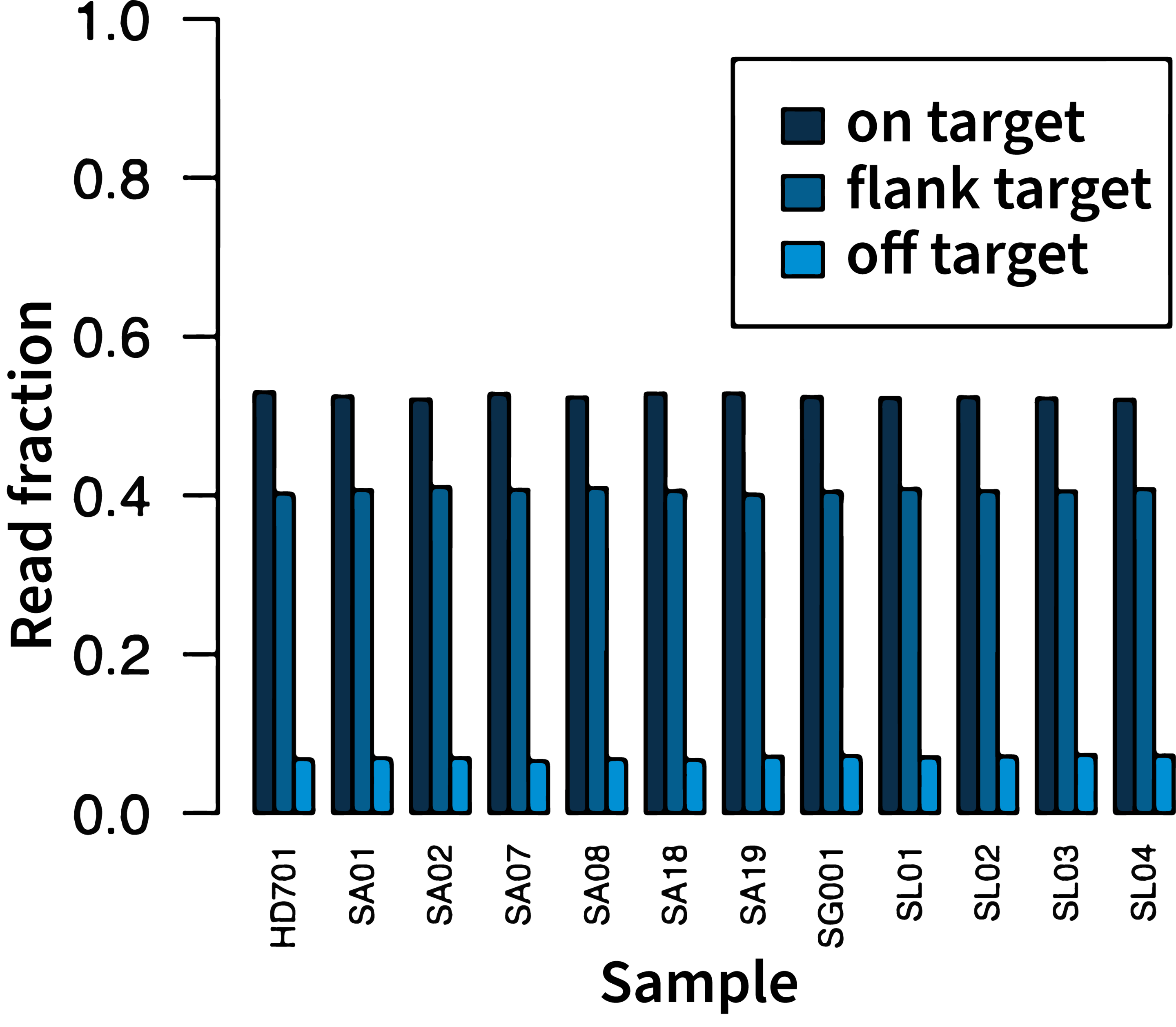
Trust your data
High on-target rate
In a typical run, the on-target rate (on-target + flanking target reads) is over 87%, maximizing the number of useful reads and increasing the multiplexing capacity.
Data on file.
Trust your data
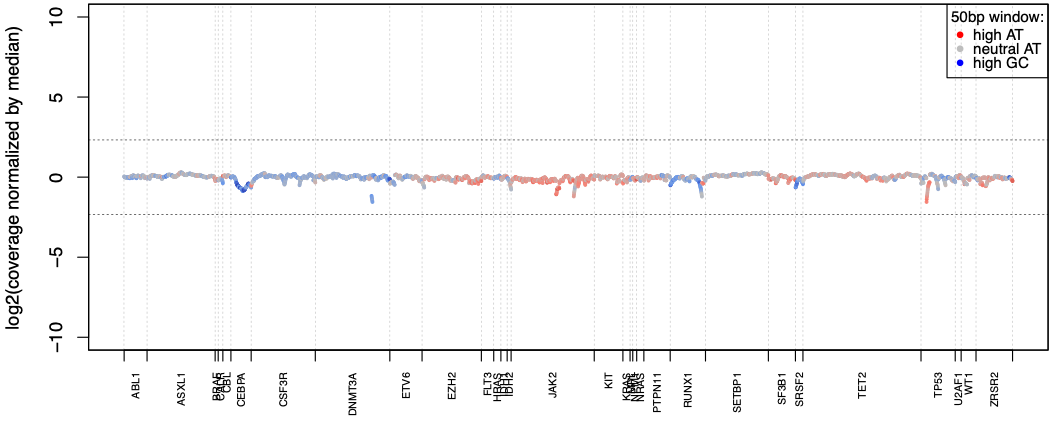
Excellent coverage uniformity
SOPHiA DDM™️ Myeloid Solution reaches over 99% coverage uniformity across AT- and GC-rich regions of the targeted genes, making each sequencing exceptionally cost-efficient.
Data on file.
Trust your data
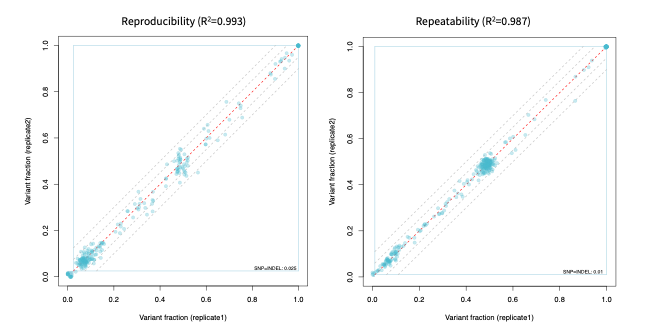
High reproducibility and repeatability
With almost 100% reproducibility and repeatability, SOPHiA DDM™️ Myeloid Solution provides consistent inter- and intra-runs results, insuring stable and trustworthy sequencing data.
Data on file.
Trust your data
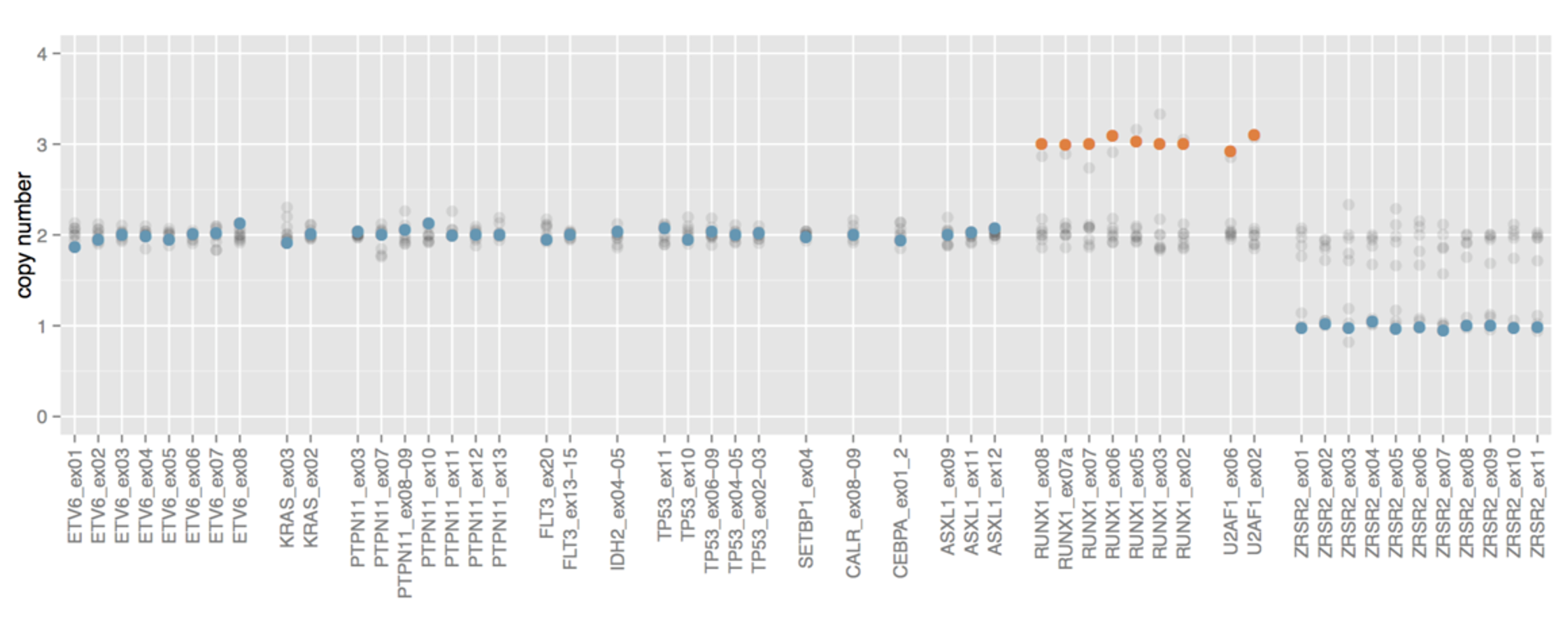
Powerful CNVs detection
SOPHiA DDM™️ Myeloid Solution is able to detect CNVs in all the genes of the panel, an important feature given their importance in tumors like AML.
Data on file.
Trust your data
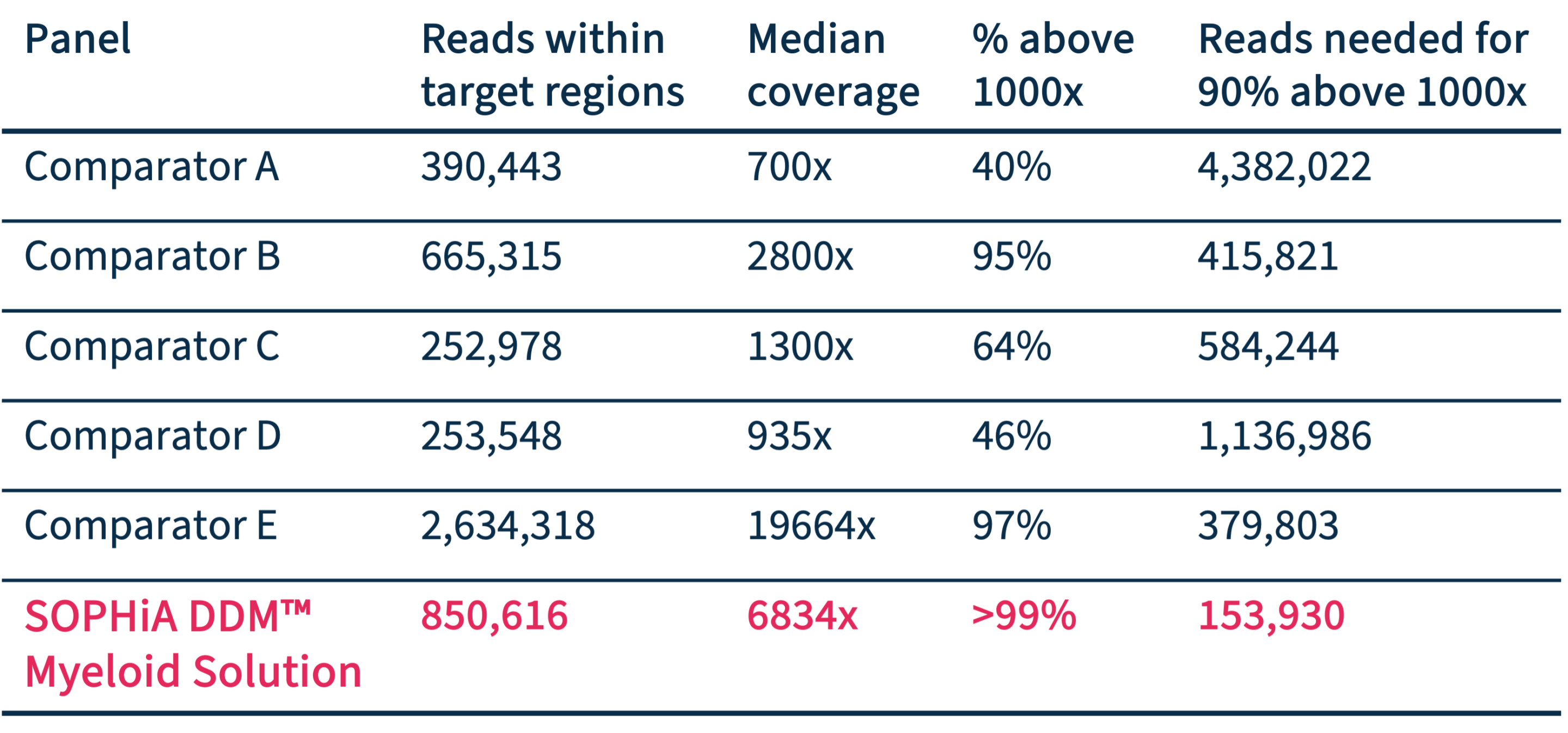
Optimal mapping performance
Compared to other amplicon-based panels, SOPHiA DDM™️ Myeloid Solution shows very good mapping to target regions, reducing the number of reads needed for the highest coverage.
Data on file.
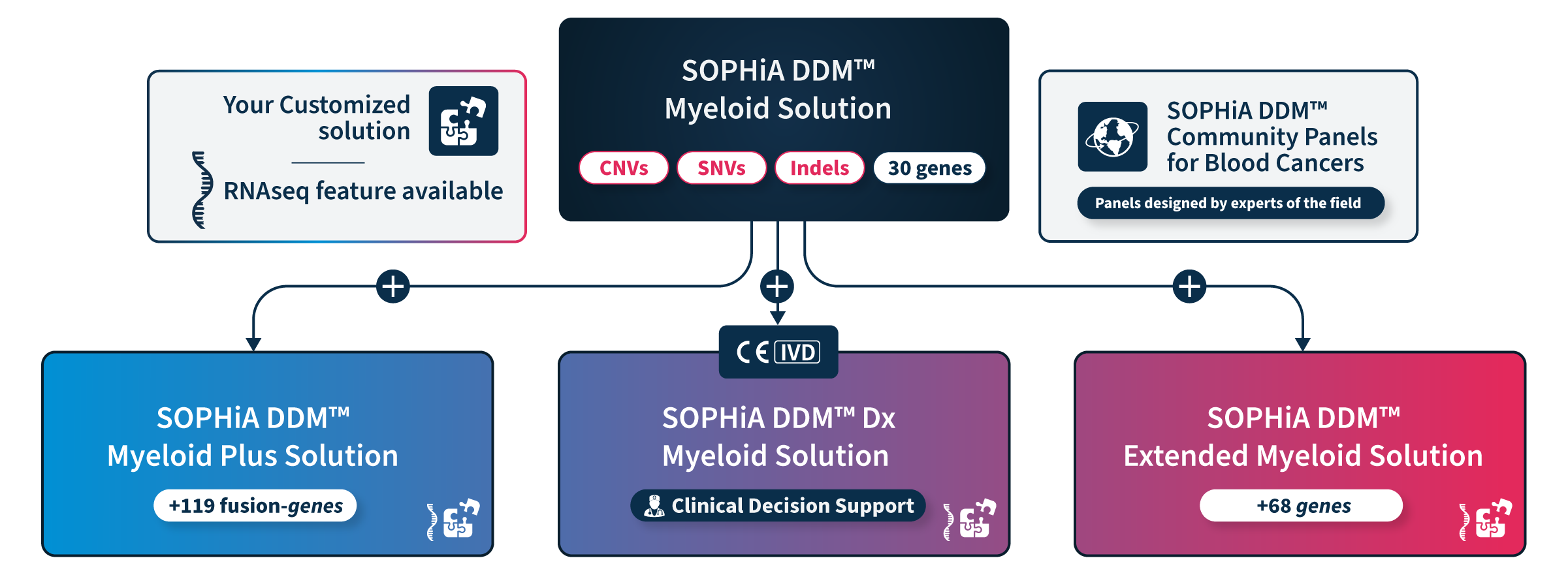
Choose the solution that fits your needs
Our comprehensive portfolio offers a range of ready-to-use and customizable solutions that enable precise characterization of myeloid malignancies driver genes by targeting key variants from blood or bone marrow samples.
All Solutions
All our solutions accurately detect SNVs, Indels, CNVs, and FLT3 internal tandem duplications (ITDs) in genes associated with myelodysplastic syndromes, myeloproliferative neoplasms, and leukemia, with additional genes and features such as the RNAseq module to answer specific needs.
SOPHiA GENETICS™ Community Panels for Blood Cancers
SOPHiA GENETICS™ Community Panels for Blood Cancers were developed and tested by SOPHiA GENETICS’s peer-network of experts in hematological malignancies to minimize set-up challenges and accelerate your workflow.
Learn directly from the experts of the field

Noah Brown, MD
Clinical Associate Professor, Roger Cancer Center, University of Michigan Health
Watch Dr Brown’s webinar to learn how he implemented a customized SOPHiA DDM™️ Myeloid Plus Solution in his academic clinical laboratory.

Mohammad Hussaini, MD
Pathologist, Moffitt Cancer Center
Listen to Dr Hussaini explain the reasons for SOPHiA DDM™️ Extended Myeloid Solution adoption at Moffitt Cancer Center.

Irina Krier, PhD
Bioinformatics Manager, Quality Expert, SOPHiA GENETICS
Let Dr Krier show you how SOPHiA DDM™️ myeloid solutions accurately detect variants in difficult genes such as CEPBA and FLT3.
Specifications
SOPHiA DDM™Myeloid Solution |
SOPHiA DDM™Myeloid Plus Solution |
SOPHiA DDM™Extended Myeloid Solution |
|
| Associated disorders | Myelodysplastic syndromes, myeloproliferative neoplasms, and leukemia | ||
| Content | 30 genes | 30 genes + 119 RNA genes fusions |
98 genes (full coding regions) |
| Key biomarkers | CEBPA, ASXL1, CALR, FLT3 -ITDs | CEBPA, ASXL1, CALR, FLT3 –ITDs, KMT2A-PTDs | |
| Target Region Size | 48 kb | 49 kb | 325 kb |
| Sample Type | Blood and bone marrow | Blood and bone marrow | Blood and bone marrow |
| Starting material | 200 ng DNA | 200 ng DNA 500 ng RNA |
200 ng DNA |
| Sequencer Compatibility |
Illumina MiSeq®, NextSeq® and Thermo Fisher Scientific Ion Proton™️ System MGI DNBSEQ-G400 |
Illumina MiSeq® and NextSeq® | Illumina NextSeq® and NovaSeq™️ |
| Library Preparation Time | 1.5 days | 1.5 days for DNA 6 hours for RNA |
1.5 days |
| Analysis time from FASTQ* | 4 hours | 4 hours | 6 hours |
| Detected Variants | SNVs Indels CNVs |
SNVs Indels CNVs Gene fusions |
SNVs Indels CNVs |
*Varies depending on the number of genes, samples multiplexed and server load.
Exons and fusions available on dedicated fact sheets.
ITDs, internal tandem duplications; PTDs, partial tandem duplications.
Resources
Fact sheets and flyers
Want to know more?
Get in touch with us.
Our client services team is on hand to help.
References
- Khoury JD, Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF, et al. The 5th edition of the World Health Organization classification of Haematolymphoid Tumours: Myeloid and histiocytic/dendritic neoplasms. Leukemia. 2022;36(7):1703–19. http://dx.doi.org/10.1038/s41375-022-01613-1
- National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology: myeloproliferative neoplasms. Version 3.2022. [Updated: Aug 2022; Accessed: Jan 2023]
- Döhner H, Wei AH, Appelbaum FR, Craddock C, DiNardo CD, Dombret H, et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood. 2022;140(12):1345–77. https://dx.doi.org/10.1182/blood.2022016867
- Smith G, Apperley J, Milojkovic D, Cross NCP, Foroni L, Byrne J, et al. A British Society for Haematology Guideline on the diagnosis and management of chronic myeloid leukaemia. Br J Haematol. 2020;191(2):171–93. http://dx.doi.org/10.1111/bjh.16971
- Gambacorti-Passerini CB, Donadoni C, Parmiani A, Pirola A, Redaelli S, Signore G, et al. Recurrent ETNK1 mutations in atypical chronic myeloid leukemia. Blood. 2015;125(3):499–503. https://dx.doi.org/10.1182/blood-2014-06-579466